Abstract
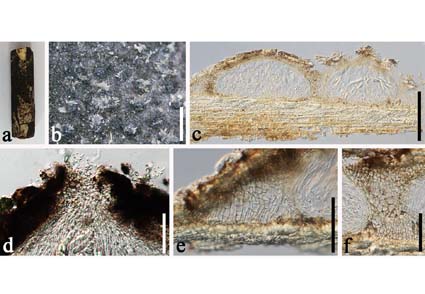
A sample of bambusicolous ascomycete was collected from Dali, Yunnan Province, China. Multi-gene phylogenetic analyses based on a combined dataset of LSU, ITS, SSU, and tef1-α sequences showed that the new collection clusters within Parabambusicola s. str. but forms a separate branch (100% ML, 1.00 PP) basal to other Parabambusicola species. Additionally, morphological characteristics of the fungus resemble the members of Parabambusicola, with 8-spored asci, and fusiform to vermiform ascospores. The species was distinguished based on morphological and phylogenetic studies, thus, it is introduced here as a new species, viz., Parabambusicola yunnanensis. Morphological description, illustrations, phylogenetic tree, and a synoptic table of morphological characteristics for Parabambusicola are provided to show the taxonomic placement of the new species.
References
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ariyawansa, H.A., Camporesi, E., Thambugala, K.M., Mapook, A., Kang, J.C., Alias, S.A., Chukeatirote, E., Thines, M., McKenzie, E.H.C. & Hyde, K.D. (2014) Confusion surrounding Didymosphaeria—phylogenetic and morphological evidence suggest Didymosphaeriaceae is not a distinct family. Phytotaxa 176: 102–119. https://doi.org/10.11646/phytotaxa.176.1.12</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ariyawansa, H.A., Hyde, K.D., Jayasiri, S.C., Buyck, B., Chethana, K.W.T., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Jayawardena, R.S., Lücking, R., Ghobad-Nejhad, M., Niskanen, T., Thambugala, M., Voigt, K., Zhao, R.L., Li, G.J., Doilom, M., Boonmee, S., Yang, Z.L., Cai, Q., Cui, Y.Y., Bahkali, A.H., Chen, J., Cui, B.K. Chen, J.J., Dayarathne, M.C., Dissanayake, A.J., Ekanayaka, A.H., Hashimoto, A., Hongsanan, S., Jones, E.B.G., Larsson, E., Li, W.J., Li, Q.R., Liu, J.K., Luo, Z.L., Maharachchikumbura, S.S.N., Mapook, A., McKenzie, E.H.C., Norphanphoun, C., Konta, S., Pang, K.L., Perera, R.H., Phookamsak, R., Phukhamsakda, C., Pinruan, U., Randrianjohany, E., Singtripop, C., Tanaka, K., Tian, C.M., Tibpromma, S., Abdel-Wahab, M.A., Wanasinghe, D.N., Wijayawardene, N.N., Zhang, J.F., Zhang, H., Abdel-Aziz, F.A., Wedin, M., Westberg, M., Ammirati, J.F., Bulgakov, T.S., Lima, D.X., Callaghan, T.M., Callac, P., Chang, C.H., Coca, L.F., Dal-Forno, M., Dollhofer, V., Fliegerová, K., Greiner, K., Griffith, G.W., Ho, H.M., Hofstetter, V., Jeewon, R., Kang, J.C., Wen, T.C., Kirk, P.M., Kytövuori, I., Lawrey, J.D., Xing, J., Li, H., Liu, Z.Y., Liu, X.Z., Liimatainen, K., Lumbsch, H.T., Matsumura, M., Moncada, B., Nuankaew, S., Parnmen, S., de Azevedo Santiago, A.L.C.M., Sommai, S., Song, Y., de Souza, C.A.F., de Souza-Motta, C.M., Su, H.Y., Suetrong, S., Wang, Y., Wei, S.F., Wen, T.C., Yuan, H.S., Zhou, L.W., Réblová, M., Fournier, J., Camporesi, E., Luangsa-ard, J.J., Tasanathai, K. & Khonsanit, A. (2015) Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 75: 27–274. https://doi.org/10.1007/s13225-015-0346-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009) trimAl: a tool for automatedalignment trimming in large-scale phylogenetic analyses. Bioinformatics 25 (15): 1972–1973. https://doi.org/10.1093/bioinformatics/btp348</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Crous, P.W., Carris, L.M., Giraldo, A., Groenewald, J.Z., Hawksworth, D.L., Hernández-Restrepo, M., Jaklitsch, W.M., Lebrun, M.H., Schumacher, R.K., Stielow, J.B., van der Linde, E.J., Vilcâne, J., Voglmayr, H. & Wood, A.R. (2015) The Genera of Fungi - fixing the application of the type species of generic names – G 2: Allantophomopsis, Latorua, Macrodiplodiopsis, Macrohilum, Milospium, Protostegia, Pyricularia, Robillarda, Rotula, Septoriella, Torula, and Wojnowicia. IMA Fungus 6: 163–198. https://doi.org/10.5598/imafungus.2015.06.01.11</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Crous, P.W., Summerell, B.A., Shivas, R.G., Romberg, M., Mel’nik, V.A., Verkley, G.J.M. & Groenewald, J.Z. (2011) Fungal Planet description sheets: 92–106. Persoonia - Molecular Phylogeny and Evolution of Fungi 27: 130–162. https://doi.org/10.3767/003158511X617561</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Crous, P.W., Wingfield, M.J., Schumacher, R.K., Summerell, B.A., Giraldo, A., Gené, J., Guarro, J., Wanasinghe, D.N., Hyde, K.D., Camporesi, E., Gareth Jones, E.B., Thambugala, K.M., Malysheva, E.F., Malysheva, V.F., Acharya, K., Álvarez, J., Alvarado, P., Assefa, A., Barnes, C.W., Bartlett, J.S., Blanchette, R.A., Burgess, T.I., Carlavilla, J.R., Coetzee, M.P.A., Damm, U., Decock, C.A. den Breeÿen, A., de Vries, B., Dutta, A.K., Holdom, D.G., Latham, S.R., Manjón, J.L., Marincowitz, S., Mirabolfathy, M., Moreno, G., Nakashima, C., Papizadeh, M., Fazeli, S.A.S., Amoozegar, M.A., Romberg, M.K., Shivas, R.G., Stalpers, J.A., Stielow, B., Stukely, M.J.C., Swart, W.J., Tan, Y.P., van der Bank, M., Wood, A.R., Zhang, Y. & Groenewald, J.Z. (2014) Fungal Planet Description Sheets: 281–319. Persoonia - Molecular Phylogeny and Evolution of Fungi 33: 212–289. https://doi.org/10.3767/003158514X685680</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dai, D.Q., Bahkali, A.H., Li, W.J., Bhat, D.J., Zhao, R.L. & Hyde, K.D. (2015) Bambusicola loculata sp. nov. (Bambusicolaceae) from bamboo. Phytotaxa 213: 122–130. https://dx.doi.org/10.11646/phytotaxa.213.2.5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dai, D.Q., Bhat, D.J., Liu, J.K., Chukeatirote, E., Zhao, R.L. & Hyde, K.D. (2012) Bambusicola, a new genus from bamboo with asexual and sexual morphs. Cryptogamie Mycologie 33: 363–379. https://doi.org/10.7872/crym.v33.iss3.2012.363</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dai, D.Q., Phookamsak, R., Wijayawardene, N.N., Li, W.J., Bhat, D.J., Xu, J.C., Taylor, J.E., Hyde, K.D. & Chukeatirote, E. (2017) Bambusicolous fungi. Fungal Diversity 82: 1–105. https://doi.org/10.1007/s13225-016-0367-8</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dai, D.Q., Wijayawardene, N.N., Dayarathne, M.C., Kumla, J., Han, L.S., Zhang, G.Q., Zhang, X., Zhang, T.T. & Chen, H.H. (2022) Taxonomic and Phylogenetic Characterizations Reveal Four New Species, Two New Asexual Morph Reports, and Six New Country Records of Bambusicolous Roussoella from China. Journal of Fungi 8: 532. https://doi.org/10.3390/jof8050532</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dong, W., Wang, B., Hyde, K.D., McKenzie, E.H.C., Raja, H.A., Tanaka, K., Abdel?Wahab, M.A., Abdel?Aziz, F.A., Doilom, M., Phookamsak, R., Hongsanan, S., Wanasinghe, D.N., Yu, X.D., Wang, G.N., Yang, H., Yang, J., Thambugala, K.M., Tian, Q., Luo, Z.L., Yang, J.B., Miller, A.N., Fournier, J., Boonmee, S., Hu, D.M., Nalumpang, S. & Zhang, H. (2020) Freshwater Dothideomycetes. Fungal Diversity 105: 319–575. https://doi.org/10.1007/s13225-020-00463-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Glez-Peña, D., Gómez-Blanco, D., Reboiro-Jato, M., Fdez-Riverola, F. & Posada, D. (2010) ALTER: program-oriented conversion of DNA and protein alignments. Nucleic Acids Research 38: 14–18. https://doi.org/10.1093/nar/gkq321</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hall, T. (2004) BioEdit. Ibis Therapeutics, Carlsbad, CA, 92008, USA. Available from: http://www.mbio.ncsu.edu/BioEdit/bioedit.html/ (accessed 18 February 2023).</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hyde, K.D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D.J., Jones, E.B.G., Liu, N.G., Abeywickrama, P.D., Mapook, A., Wei, D.P., Perera, R.H., Manawasinghe, I.S., Pem, D., Bundhun, D., Karunarathna, A., Ekanayaka, A.H., Bao, D.F., Li, J.F., Samarakoon, M.C., Chaiwan, N., Lin, C.G., Phutthacharoen, K., Zhang, S.N., Senanayake, I.C., Goonasekara, I.D., Thambugala, K.M., Phukhamsakda, C., Tennakoon, D.S., Jiang, H.B., Yang, J., Zeng, M., Huanraluek, N., Liu, J.K., Wijesinghe, S.N., Tian, Q., Tibpromma, S., Brahmanage, R.S., Boonmee, S., Huang, S.K., Thiyagaraja, V., Lu, Y.Z., Jayawardena, R.S., Dong, W., Yang, E.F., Singh, S.K., Singh, S.M., Rana, S., Lad, S.S., Anand, G., Devadatha, B., Niranjan, M., Sarma, V.V., Liimatainen, K., Aguirre?Hudson, B., Niskanen, T., Overall, A., Alvarenga, R.L.M., Gibertoni, T.B., Pfliegler, W.P., Horvaìth, E., Imre, A., Alves, A.L., Santos, A.C.S., Tiago, P.V., Bulgakov, T.S., Wanasinghe, D.N., Bahkali, A.H., Doilom, M., Elgorban, A.M., Maharachchikumbura, S.S.N., Rajeshkumar, K.C., Haelewaters, D., Mortimer, P.E., Zhao, Q., Lumyong, S., Xu, J.C. & Sheng, J. (2020) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. https://doi.org/10.1007/s13225-020-00439-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hirayama, K., Tanaka, K., Raja, H.A., Miller, A.N. & Shearer, C.A. (2010) A molecular phylogenetic assessment of Massarina ingoldiana sensu lato. Mycologia 102: 729–747. https://doi.org/10.3852/09-230</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Index Fungorum (2022) Available from: http://www.indexfungorum.org/names/IndexFungorumRegisterName.asp (accessed 18 February 2022).</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Jayasiri, S.C., Hyde, K.D., Ariyawansa, H.A., Bhat, J.D., Buyck, B., Cai, L., Dai, Y.C., Abd-Elsalam, K.A., Ertz, D., Hidayat, I., Jeewon, R., Jones, E.B.G., Bahkali, A.H., Karunarathna, S.C., Liu, J.K., Luangsa-ard, J.J., Lumbsch, H.T., Maharachchikumbura, S.S.N., McKenzie, E.H.C., Moncalvo, J.M., Ghobad-Nejhad, M., Nilsson, H., Pang, K.A., Pereira, O.L., Phillips, A.J.L., Raspé, O., Rollins, A.W., Romero, A.I., Etayo, J., Selçuk, F., Stephenson, S.L., Suetrong, S., Taylor, J.E., Tsui, C.K.M., Vizzini, A., Abdel-Wahab, M.A., Wen, T.C., Boonmee, S., Dai, D.Q., Daranagama, D.A., Dissanayake, A.J., Ekanayaka, A.H., Fryar, S.C., Hongsanan, S., Jayawardena, R.S., Li, W.J., Perera, R.H., Phookamsak, R., de Silva, N.I., Thambugala, K.M., Tian, Q., Wijayawarden, N.N., Zhao, R.L., Zhao, Q., Kang, J.C. & Promputtha, I. (2015) The Faces of fungi database: fungal names linked with morphology and phylogeny and human impacts. Fungal Diversity 74: 3–18. https://doi.org/10.1007/s13225-015-0351-8</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kruys, A., Eriksson, O.E. & Wedin, M. (2006) Phylogenetic relationships of coprophilous Pleosporales (Dothideomycetes, Ascomycota), and the classification of some bitunicate taxa of unknown position. Mycological Research 110: 527–536. https://doi.org/10.1016/j.mycres.2006.03.002</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, J.K., Hyde, K.D., Jones, E.B.G., Ariyawansa, H.A., Bhat, D.J., Boonmee, S., Maharachchikumbura, S.S.N., McKenzie, E.H.C., Phookamsak, R., Phukhamsakda, C., Shenoy, B.D., Abdel-Wahab, M.A., Buyck, B., Chen, J., Chethana, K.W.T., Singtripop, C., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Dissanayake, A.J., Doilom, M., D’souza, M.J., Fan, X.L., Goonasekara, I.D., Hirayama, K., Hongsanan, S., Jayasiri, S.C., Jayawardena, R.S., Karunarathna, S.C., Li, W.J., Mapook, A., Norphanphoun, C., Pang, K.L., Perera, R.H., Peršoh, D., Pinruan, U., Senanayake, I.C., Somrithipol, S., Suetrong, S., Tanaka, K., Thambugala, K.M., Tian, Q., Tibpromma, S., Udayanga, D., Wijayawardene, N.N., Wanasinghe, D., Wisitrassameewong, K., Zeng, X.Y., Abdel-Aziz, F.A., Adam?ík, S., Bahkali, A.H., Boonyuen, N., Bulgakov, T., Callac, P., Chomnunti, P., Greiner, K., Hashimoto, A., Hofstetter, V., Kang, J.C., Lewis, D., Li, X.H., Liu, X.Z., Liu, Z.Y., Matsumura, M., Mortimer, P.E., Rambold, G., Randrianjohany, E., Sato, G., Sri-Indrasutdhi, V., Tian, C.M., Verbeken, A., von Brackel, W., Wang, Y., Wen, T.C., Xu, J.C., Yan, J.Y., Zhao, R.L. & Camporesi, E. (2015) Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. https://doi.org/10.1007/s13225-015-0324-y</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Li, G.J., Hyde, K.D., Zhao, R.L., Hongsanan, S., Abdel-Aziz, F.A., Abdel-Wahab, M.A., Alvarado, P., Alves-Silva, G., Ammirati, J.F., Ariyawansa, H.A., Baghela, A., Bahkali, A.H., Beug, M., Bhat, D.J., Bojantchev, D., Boonpratuang, T., Bulgakov, T.S., Camporesi, E., Boro, M.C., Ceska, O., Chakraborty, D., Chen, J.J., Chethana, K.W.T., Chomnunti, P., Consiglio, G., Cui, B.K., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Das, K., Dayarathne, M.C., De Crop, E., De Oliveira, R.J.V., de Souza, C.A.F., de Souza, J.I., Dentinger, B.T.M., Dissanayake, A.J., Doilom, M., Drechsler-Santos, E.R., Ghobad-Nejhad, M., Gilmore, S.P., Góes-Neto, A., Gorczak, M., Haitjema, C.H., Hapuarachchi, K.K., Hashimoto, A., He, M.Q., Henske, J.K., Hirayama, K., Iribarren, M.J., Jayasiri, S.C., Jayawardena, R.S., Jeon, S.J., Jerônimo, G.H., Jesus, A.L., Jones, E.B.G., Kang, J.C., Karunarathna, S.C., Kirk, P.M., Konta, S., Kuhnert, E., Langer, E., Lee, H.S., Lee, H.B., Li, W.J., Li, X.H., Liimatainen, K., Lima, D.X., Lin, C.G., Liu, J.K., Liu, X.Z., Liu, Z.Y., Luangsa-ard, J.J., Lücking, R., Lumbsch, H.T., Lumyong, S., Leaño, E.M., Marano, A.V., Matsumura, M., McKenzie, E.H.C., Mongkolsamrit, S., Mortimer, P.E., Nguyen, T.T.T., Niskanen, T., Norphanphoun, C., O’Malley, MA., Parnmen, S., Paw?owska, J., Perera, R.H., Phookamsak, R., Phukhamsakda, C., Pires-Zottarelli, C.L.A., Raspé, O., Reck, M.A., Rocha, S.C.O., de Santiago, A.L.C.M.A., Senanayake, I.C., Setti, L., Shang, Q.J. & Singh, SK. (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78: 1–237. https://doi.org/10.1007/s13225-016-0366-9</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, J.K., Hyde, K.D., Jeewon, R., Phillips, A.J.L., Maharachchikumbura, S.S.N., Ryberg, M., Liu, Z.Y. & Zhao, Q. (2017) Ranking higher taxa using divergence times: a case study in Dothideomycetes. Fungal Diversity 84: 75–99. https://doi.org/10.1007/s13225-017-0385-1</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, N.G., Hongsanan, S., Yang, J., Lin, C.G., Bhat, D.J., Liu, J.K., Jumpathong, J., Boonmee, S., Hyde, K.D. & Liu, Z.Y. (2017) Dendryphiella fasciculata sp. nov. and notes on other Dendryphiella species. Mycosphere 8: 1575–1586. https://doi.org/10.5943/mycosphere/8/9/12</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ma, X. (2016) Study on Complete Mitochondrial Genome of Cypridopsis vidua and Molecular Phylogeny of Ostracoda. Ph.D. Thesis, East China Normal University, Shanghai, China.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop 2010: 1–8. https://doi.org/10.1109/GCE.2010.5676129</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Phookamsak, R., Manamgoda, D.S., Li, W.J., Dai, D.Q., Singtripop, C. & Hyde, K.D. (2015) Poaceascoma helicoides gen et sp. nov., a new genus with scolecospores in Lentitheciaceae. Cryptogamie, Mycologie 36: 225–236. https://doi.org/10.7872/crym/v36.iss2.2015.225</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Phookamsak, R., Hyde, K.D., Jeewon, R., Bhat, D.J., Jones, E.B.G., Maharachchikumbura, S.S.N., Raspé, O., Karunarathna, S.C., Wanasinghe, D.N., Hongsanan, S., Doilom, M., Tennakoon, D.S., Machado, A.R., Firmino, A.L., Ghosh, A., Karunarathna, A., Meši?, A., Dutta, A.K., Thongbai, B., Devadatha, B., Norphanphoun, C., Senwanna, C., Wei, D., Pem, D., Ackah, F.K., Wang, G.N., Jiang, H.B., Madrid, H., Lee, H.B., Goonasekara, I.D., Manawasinghe, I.S., Kušan, I., Cano, J., Gené, J., Li, J.f., Das, K., Acharya, K., Raj, K.N.A., Latha, K.P.D., Chethana, K.W.T., He, M.Q., Dueñas, M., Jadan, M., Martín, M.P., Samarakoon, M.C., Dayarathne, M.C., Raza, M., Park, M.S., Telleria, M.T., Chaiwan, N., Mato?ec, N., de Silva, N.I., Pereira, O.L., Singh, P.N., Manimohan, P., Uniyal, P., Shang, Q.J., Bhatt, R.P., Perera, R.H., Alvarenga, R.L.M., Nogal-Prata, S., Singh, S.K., Vadthanarat, S., Oh, S.Y., Huang, S.K., Rana, S., Konta, S., Paloi, S., Jayasiri, S.C., Jeon, S.J., Mehmood, T., Gibertoni, T.B., Nguyen, T.T.T., Singh, U., Thiyagaraja, V., Sarma, V.V., Dong, W., Yu, X.D., Lu, Y.Z., Lim, Y.W., Chen, Y., Tkal?ec, Z., Zhang, Z.F., Luo, Z.L., Daranagama, D.A., Thambugala, K.M., Tibpromma, S., Camporesi, E., Bulgakov, T.S., Dissanayake, A.J., Senanayake, I.C., Dai, D.Q., Tang, L.Z., Khan, S., Zhang, H., Promputtha, I., Cai, L., Chomnunti, P., Zhao, R.L., Lumyong, S., Boonmee, S., Wen, T.C., Mortimer, P.E. & Xu, J.C. (2019) Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Diversity 95: 1–273. https://doi.org/10.1007/s13225-019-00421-w</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Phookamsak, R., Jiang, H.B., Suwannarach, N., Lumyong, S., Xu, J.C., Xu, S., Liao, C.F. & Chomnunti, P. (2022) Bambusicolous Fungi in Pleosporales: Introducing Four Novel Taxa and a New Habitat Record for Anastomitrabeculia didymospora. Journal of Fungi 8 (6): 630. https://doi.org/10.3390/jof8060630</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Phukhamsakda, C., Bhat, D.J., Hongsanan, S., Xu, J.C., Stadler, M. & Hyde, K.D. (2018) Two novel species of Neoaquastroma (Parabambusicolaceae, Pleosporales) with their phoma-like asexual morphs. MycoKeys 34: 47–62. https://doi.org/10.3897/mycokeys.34.25124</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Phukhamsakda, C., McKenzie, E.H.C., Phillips, A.J.L., Jones, E.B.G., Bhat, D.J., Stadler, M., Bhunjun, C.S., Wanasinghe, D.N., Thongbai, B., Camporesi, E., Ertz, D., Jayawardena, R.S., Perera, R.H., Ekanayake, A.H., Tibpromma, S., Doilom, M., Xu, J.C. & Hyde, K.D. (2020) Microfungi associated with Clematis (Ranunculaceae) with an integrated approach to delimiting species boundaries. Fungal diversity 102: 1–203. https://doi.org/10.1007/s13225-020-00448-4</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Quaedvlieg, W., Verkley, G.J.M., Shin, H.D., Barreto, R.W., Alfenas, A.C., Swart, W.J., Groenewald, J.Z. & Crous, P.W. (2013) Sizing up Septoria. Studies in Mycology 75: 307–390. https://doi.org/10.3114/sim0017</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rambaut, A. (2012) FigTree version 1.4, 2, University of Edinburgh Edinburgh.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rannala, B. & Yang, Z. (1996) Probability distribution of molecular evolutionary trees, a new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. https://doi.org/10 1007/BF02338839 </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rehner, S. (2001) Primers for Elongation Factor 1-alpha (EF1-alpha) Insect Biocontrol Laboratory: USDA, ARS, PSI. [rehner@ba.ars. usda.gov]</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ronquist, F., Huelsenbeck, J. & Teslenko, M. (2011) Draft MrBayes version 3.2. Manual, tutorials and model summaries. Bioinformatics 85–131. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Samarakoon, M.C., Wanasinghe, D.N., Liu, J.K., Hyde, K.D. & Promputtha, I. (2019) The genus Neoaquastroma is widely distributed., a taxonomic novelty, N. cylindricum sp. nov. (Parabambusicolaceae, Pleosporales) from Guizhou, China. Asian Journal of Mycology 2: 235–244. https://doi.org/10.5943/ajom/2/1/14</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Senanayake, I.C., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S.N., Bundhun, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J. & Xiang, M.M. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 678–2754. https://doi.org/10.5943/mycosphere/11/1/20</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Schoch, C.L., Crous, P.W., Groenewald, J.Z., Boehm, E.W.A., Burgess, T.I., de Gruyter, J., de Hoog, G.S., Dixon, L.J., Grube, M., Gueidan, C., Harada, Y., Hatakeyama, S., Hirayama, K., Hosoya, T., Huhndorf, S.M., Hyde, K.D., Jones, E.B.G., Kohlmeyer, J., Kruys, Å., Li, Y.M., Lücking, R., Lumbsch, H.T., Marvanová, L., Mbatchou, J.S., McVay, A.H., Miller, A.N., Mugambi, G.K., Muggia, L., Nelsen, M.P., Nelson, P., Owensby, C.A., Phillips, A.J.L., Phongpaichit, S., Pointing, S.B., Pujade-Renaud, V., Raja, H.A., Plata, E.R., Robbertse, B., Ruibal, C., Sakayaroj, J., Sano, T., Selbmann, L., Shearer, C.A., Shirouzu, T., Slippers, B., Suetrong, S., Tanaka, K., Volkmann-Kohlmeyer, B., Wingfield, M.J., Wood, A.R., Woudenberg, J.H.C., Yonezawa, H., Zhang, Y. & Spatafora, J.W. (2009) A class-wide phylogenetic assessment of Dothideomycetes. Studies in Mycology 64: 1–15. https://doi.org/10.3114/sim.2009.64.01</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stamatakis, A., Hoover, P. & Rougemont, J. (2008) A rapid bootstrap algorithm for the ML web servers. Systematic biology 57: 758–771. https://doi.org/10 1080/10635150802429642 </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9): 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Sung, G.H., Hywel-Jones, N.L., Sung, J.M., Luangsa-Ard, J.J., Shrestha, B. & Spatafora, J.W. (2007) Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Studies in Mycology 57: 5–59. https://doi.org/10.3114/sim.2007.57.01</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tanaka, K. & Harada, Y. (2003) Pleosporales in Japan (3). The genus Massarina. Mycoscience 44: 173–185. https://doi.org/10.1007/S10267-003-0102-7</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tanaka, K., Hirayama, K., Yonezawa, H., Sato, G., Toriyabe, A., Kudo, H., Hashimoto, A., Matsumura, M., Harada, Y., Kurihara, Y., Shirouzu, T. & Hosoya, T. (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes) Studies in Mycology 82: 75–136. https://doi.org/10.1016/j.simyco.2015.10.002</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tsui, C.K.M., Berbee, M.L., Jeewon, R. & Hyde, K.D. (2006) Molecular phylogeny of Dictyosporium and allied genera inferred from ribosomal DNA. Fungal Diversity 21: 157–166.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Verkley, G.J.M., da Silva, M., Wicklow, D.T. & Crous, P.W. (2004) Paraconiothyrium, a new genus to accommodate the mycoparasite Coniothyrium minitans, anamorphs of Paraphaeosphaeria, and four new species. Studies in Mycology 50: 323–336.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/JB.172.8.4238-4246</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wanasinghe, D.N., Hyde, K.D., Konta, S., To-Anun, C. & Jones, E.B.G. (2017) Saprobic Dothideomycetes in Thailand: Neoaquastroma gen. nov. (Parabambusicolaceae) introduced based on morphological and molecular data. Phytotaxa 302: 133–144. https://doi.org/10.11646/phytotaxa.302.2.3</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wijayawardene, N.N., Dai, D.Q., Zhu, M.L., Wanasinghe, D.N., Kumla, J., Zhang, G.Q., Zhang, T.T., Han, L.S., Tibpromma, S. & Chen, H.H. (2022) Fungi associated with dead branches of Magnolia grandiflora: A case study from Qujing, China. Frontiers in Microbiology 13. https://doi.org/10.3389/fmicb.2022.954680 </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wijayawardene, N.N., Hyde, K.D., Dai, D.Q., Sanchez-Garcia, M., Goto, B.T., Saxena, R.K., Erdogdu, M., Selçuk, F., Rajeshkumar, K.C., Aptroot, A., B?aszkowski, J., Boonyuen, N., da Silva, G.A., de Souza, F.A., Dong, W., Ertz, D., Haelewaters, D., Jones, E.B.G., Karunarathna, S.C., Kirk, P.M., Kukwa, M., Kumla, J., Leontyev, D.V., Lumbsch, H.T., Maharachchikumbura, S.S.N., Marguno, F., Martínez-Rodríguez, P., Meši?, A., Monteiro, J.S., Oehl, F., Paw?owska, J., Pem, D., Pfliegler, W.P., Phillips, A.J.L., Pošta, A., He, M.Q., Li, J.X., Raza, M., Sruthi, O.P., Suetrong, S., Suwannarach, N., Tedersoo, L., Thiyagaraja, V., Tibpromma, S., Tkal?ec, Z., Tokarev, Y.S., Wanasinghe, D.N., Wijesundara, D.S.A., Wimalaseana, S.D.M.K., Madrid, H., Zhang, G.Q., Gao, Y., Sánchez-Castro., Tang, L.Z., Stadler, M., Yurkov, A. & Thines, M. (2022) Outline of Fungi and fungus-like taxa. Mycosphere 13: 55–453. https://doi.org/10.5943/mycosphere/13/1/2</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Xie, N., Phookamsak, R., Jiang, H., Zeng, Y.J., Zhang, H., Xu, F., Lumyong, S., Xu, J. & Hongsanan, S. (2022) Morpho-Molecular Characterization of five novel taxa in Parabambusicolaceae (Massarineae, Pleosporales) from Yunnan, China. Journal of Fungi 8: 108. https://doi.org/10.3390/jof8020108</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Yang, E.F., Tibpromma, S., Karunarathna, S.C., Phookamsak, R., Xu, J.C., Zhao, Z.Z.X., Karunanayake, C. & Promputtha, I. (2022) Taxonomy and phylogeny of novel and extant taxa in Pleosporales associated with Mangifera indica from Yunnan, China (Series I). Journal of Fungi 8: 152. https://doi.org/10.3390/jof8020152</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Yasanthika, E., Dissanayake, L.S., Wanasinghe, D.N., Karunarathna, S.C., Mortimer, P.E., Samarakoon, B.C., Monkai, J. & Hyde, K.D. (2020) Lonicericola fuyuanensis (Parabambusicolaceae) a new terrestrial pleosporalean ascomycete from Yunnan Province, China. Phytotaxa 446: 103–113. https://doi.org/10.11646/PHYTOTAXA.446.2.3</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, Y., Schoch, C.L., Fournier, J., Crous, P.W., de Gruyter, J., Woudenberg, J.H.C., Hirayama, K., Tanaka, K., Pointing, S.B., Spatafora, J.W. & Hyde, K.D. (2009) Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Studies in Mycology 64: 85–102. https://doi.org/10.3114/sim.2009.64.04</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhaxybayeva, O. & Gogarten, J.P. (2002) Bootstrap Bayesian probability and maximum likelihood mapping, exploring new tools for comparative genome analyses. MBC genomics 3: 1–4. https://doi.org/10.1186/1471-2164-3-4</span></span></span>