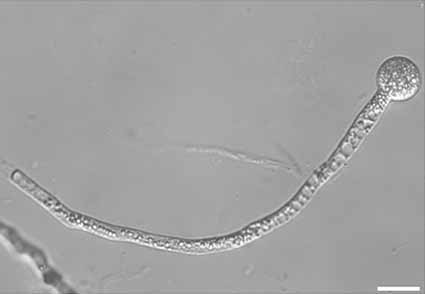
Abstract
Neoconidiobolus pseudothromboides is proposed as a species new on the basis of morphological characteristics, physiological feature and molecular phylogeny. This new species is morphologically allied to N. thromboides. However, the maximum growth temperature of this new species is lower than that of N. thromboides and it grows slower than N. thromboides. The phylogenetic analysis of mitochondrial small subunit (mtSSU), nuclear large subunit (nucLSU) and translation elongation-factor-like (EFL) reveals that Neoconidiobolus is divided into three clades, and N. pseudothromboides sp. nov. is closely related to N. lachnodes rather than N. thromboides. Morphological comparisons between N. pseudothromboides sp. nov. and its affinities are provided herein.
References
Chen, C., Ye, S.D., Wang, D.Q., Hatting, J.L. & Yu, X.P. (2014) Alginate embedding and subsequent sporulation of in vitro-produced Conidiobolus thromboides hyphae using a pressurised air-extrusion method. Biological Control 69: 52–58. https://doi.org/10.1016/j.biocontrol.2013.10.016
Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 9 (8): 772. https://doi.org/10.1038/nmeth.2109
Drechsler, C. (1952) Widespread distribution of Delacroixia coronata and other saprophytic Entomophthoraceae in plant detritus. Science 115: 575–576. https://doi.org/10.1126/science.115.2995.575
Drechsler, C. (1953) Three new species of Conidiobolus isolated from leaf mold. Journal of the Washington Academy of Science 43 (2): 29–34.
Drechsler, C. (1954) Two species of Conidiobolus with minutely ridged zygospores. American Journal of Botany 41: 567–575. https://doi.org/10.1002/j.1537-2197.1954.tb14380.x
Drechsler, C. (1955) Three new species of Conidiobolus isolated from decaying plant detritus. American Journal of Botany 42 (5): 437–443. https://doi.org/10.1002/j.1537-2197.1955.tb11144.x
Drechsler, C. (1960) Two new species of Conidiobolus found in plantdetritus. American Journal of Botany 47: 368–377. https://doi.org/10.1002/j.1537-2197.1960.tb07138.x
Drechsler, C. (1965) A robust Conidiobolus with zygospores containing granular parietal protoplasm. Mycologia 57 (6): 913–926. https://doi.org/10.2307/3756891
Edgar, R.C. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. https://doi.org/10.1093/nar/gkh340
Gryganskyi, A.P., Humber, R.A., Smith, M.E., Hodge, K., Huang, B., Voigt, K. & Vilgalys, R. (2013) Phylogenetic lineages in Entomophthoromycota. Persoonia 30: 94–105. https://doi.org/10.3767/003158513X666330
Guindon, S. & Gascuel, O. (2003) A simple, fast and accurate method to estimate large phylogenies by maximum-likelihood. Systematic Biology 52: 696–704. https://doi.org/10.1080/10635150390235520
Hall, T.A. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Humber, R.A. (2012) Entomophthoromycota: a new phylum and reclassification for entomophthoroid fungi. Mycotaxon 120: 477–492. https://doi.org/10.5248/120.477
King, D.S. (1976) Systematics of Conidiobolus (Entomophthorales) usingnumerical taxonomy I. Taxonomic considerations. Canadian Journal of Botany 54: 45–65. https://doi.org/10.1139/b76-008
King, D.S. (1977) Systematics of Conidiobolus (Entomophthorales) using numerical taxonomy III. Descriptions of recognized species. Canadian Journal of Botany 55:718–729. https://doi.org/10.1139/b77-086
Michal, H., Ryszard, M. & Jacek, P.T. (2014) The effect of Cry1AB insecticidal protein on the incidence of entomopathogenic fungi infecting aphids on Bt maize. Zemdirbyste-agriculture 101 (3): 279–284. https://doi.org/10.13080/z-a.2014.101.036
Nie, Y., Yu, C.Z., Liu, X.Y. & Huang, B. (2012) A new species of Conidiobolus (Ancylistaceae) from Anhui, China. Mycotaxon 120: 427–435. https://doi.org/10.5248/120.427
Nie, Y., Tang, X.X., Liu, X.Y. & Huang, B. (2016) Conidiobolus stilbeus, a new species with mycelial strand and two types of primary conidiophores. Mycosphere 7 (6): 801–809. https://doi.org/10.5943/mycosphere/7/6/11
Nie, Y., Tang, X.X., Liu, X.Y. & Huang, B. (2017) A new species of Conidiobolus with chlamdosporus from Dabie Mountains, eastern China. Mycosphere 8 (7): 809–816. https://doi.org/10.5943/mycosphere/8/7/1
Nie, Y., Qin, L., Yu, D.S., Liu, X.Y. & Huang, B. (2018) Two new species of Conidiobolus occurring in Anhui, China. Mycological Progress 17 (10): 1203–1211. https://doi.org/10.1007/s11557-018-1436-z
Nie, Y., Yu, D.S., Wang, C.F., Liu, X.Y. & Huang, B. (2020a) A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): four clades including three new genera. Mycokeys 66: 55–81. https://doi.org/10.3897/mycokeys.66.46575
Nie, Y., Cai, Y., Gao, Y., Yu, D.S., Wang, Z.M., Liu, X.Y. & Huang, B. (2020b) Three new species of Conidiobolus sensu stricto from plant debris in eastern China. MycoKeys 73: 133–149. https://doi.org/10.3897/mycokeys.73.56905
Nie, Y., Wang, Z.M., Liu, X.Y. & Huang, B. (2021) A morphological and molecular survey of Neoconidiobolus reveals a new species and two new combinations. Mycological Porgress 20 (10): 1233–1241. https://doi.org/10.1007/s11557-021-01720-w
Nie, Y., Zhao, H., Wang, Z.M., Zhou, Z.Y., Liu, X.Y. & Huang, B. (2022) Two new species in Capillidium (Ancylistaceae, Entomophthorales) from China, with a proposal for a new combination. Mycokeys 89: 139–153. https://doi.org/10.3897/mycokeys.89.79537
Rambaut, A. (2012) FigTree version 1.4.0. Available at: http://tree.bio.ed.ac.uk/software/figtree/ (accessed 28 November 2022)
Ronquist, F. & Huelsenbeck, J.P. (2003) MRBAYES 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Srinivasan, M.C. & Thirumalachar, M.J. (1962) Studies on species of Conidiobolus from India-II. Sydowia, Annales Mycologici 16: 60–66
Srinivasan, M.C. & Thirumalachar, M.J. (1968) Two new species of Conidiobolus from India. Journal of the Mitchell Society 84: 211–212
Stamatakis, A. (2014) RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 (9): 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Swofford, D.L. (2002) PAUP*: Phylogenetic analysis using parsimony (*and other methods), Version 4.0b10. Sinauer Associates, Sunderland
Vaidya, G., Lohman, D.J. & Meier, R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27 (2): 171–180. https://doi.org/10.1111/j.1096-0031.2010.00329.x
Vilela, R., Silva, S.M.S., Correa, F.R., Dominguez, E. & Mendoza, L. (2010) Morphologic and phylogenetic characterization of Conidiobolus lamprauges recovered from infected sheep. Journal of Clinical Microbiology 48: 427–432. https://doi.org/10.1128/JCM.01589-09
Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Waingankar, V.M., Singh, S.K. & Srinivasan, M.C. (2008) A new thermophilic species of Conidiobolus from India. Mycopathologia 165: 173–177. https://doi.org/10.1007/s11046-007-9088-6
Watanabe, M., Lee, K., Goto, K., Kumagai, S., Sugita-Konishi, Y. & Hara-Kudo, Y. (2010) Rapid and effective DNA extraction method with bead grinding for a large amount of fungal DNA. Journal of Food Protection 73 (6): 1077–1084. https://doi.org/10.4315/0362-028X-73.6.1077
Waters, S.D. & Callaghan, A.A. (1989) Conidiobolus iuxtagenitus, a new species with discharge delongate repetitional conidia and conjugation tubes. Mycological Research 93: 223–226. https://doi.org/10.1016/S0953-7562(89)80121-2
Zhao, H., Nie, Y., Zong, T.K., Dai, Y.C. & Liu, X.Y. (2022) Three New Species of Absidia (Mucoromycota) from China Based on Phylogeny, Morphology and Physiology. Diversity 14 (2): 132. https://doi.org/10.3390/d14020132
Zheng, R.Y., Chen, G.Q., Huang, H. & Liu, X.Y. (2007) A monograph of Rhizopus. Sydowia 59: 273.
Zheng, R.Y. & Liu, X.Y. (2009) Taxa of Pilaira (Mucorales, Zygomycota) from China. Nova Hedwig 88: 255–267. https://doi.org/10.1127/0029-5035/2009/0088-0255
Zheng, R.Y., Liu, X.Y. & Li, R.Y. (2009) More Rhizomucor causing human mucormycosis from China: R. chlamydosporus sp. nov. Sydowia 61: 135–147.
Zoller, S., Scheideggera, C. & Sperisena, C. (1999) PCR primers for the amplification of mitochondrial small subunit ribosomal DNA of lichen-forming ascomycetes. Lichenologist 31 (5): 511–516. https://doi.org/10.1006/lich.1999.0220