Abstract
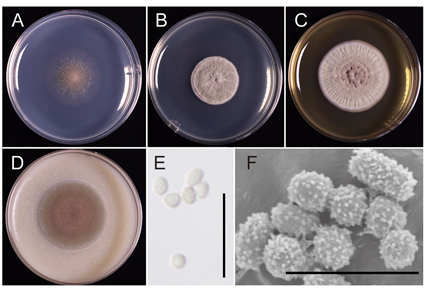
Four strains of the fungal genus Oidiodendron were isolated from the fine roots of Vaccinium boniense which is distributed only on the Bonin Islands, Japan. Two new species, Oidiodendron tokumasui (NBRC 115411T) and Oidiodendron boninense (NBRC 115392T), are described in this paper. Molecular phylogenetic analyses based on multigene sequence-data showed that each novel species formed a unique clade. Oidiodendron tokumasui is phylogenetically related to O. maius but can be distinguished from O. maius by good growth at 25‒30 ˚C and pH 7‒9, colony diameter after 28 days, and length of the conidiophores. Oidiodendron boninense is phylogenetically related to O. setiferum, O. rammosum, O. muniellense, and O. fuscum but differs from those four species in the following aspects: good growth at 25 ˚C and pH 7, absence of melanized appendages, spinulate and thick-walled conidia, and length of the conidiophores. Both new species formed ericoid mycorrhizal-like hyphal coils in the rhizodermal cells of sterile blueberry seedlings in vitro.
References
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Baba, T. & Hirose, D. (2020) Morphological characteristics of rhizodermal colonization by Leohumicola species in an ericaceous host. Plant Roots 14: 1–10. https://doi.org/10.3117/plantroot.14.1</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Baba, T. & Hirose, D. (2021) Slow-growing fungi belonging to the unnamed lineage in Chaetothyriomycetidae form hyphal coils in vital ericaceous rhizodermal cells in vitro. Fungal Biology 125: 1026–1035. https://doi.org/10.1016/j.funbio.2021.07.003</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Baba, T., Hirose, D., Noma, S. & Ban, T. (2021) Inoculation with two Oidiodendron maius strains differentially alters the morphological characteristics of fibrous and pioneer roots of Vaccinium virgatum ‘Tifblue’ cuttings. Scientia Horticulturae 281: 109948. https://doi.org/10.1016/j.scienta.2021.109948</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Couture, M., Fortin, J. & Dalpé, Y. (1983) Oidiodendron griseum Robak: an endophyte of ericoid mycorrhiza in Vaccinium spp. New Phytologist 95: 375–380. https://doi.org/10.1111/j.1469-8137.1983.tb03505.x</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Crous, P.W., Wingfield, M.J., Burgess, T.I., Hardy, G.E.S.J., Gené, J., Guarro, J., Baseia, I.G., Garcia, D., Gusmão, L.F.P., Souza-Motta, C.M., Thangavel, R., Adamik, S., Barili, A., Barnes, C.W., Bezerra, J.D.P., Bordallo, J.J., Cano-Lira, J.F., de Oliveira, R.J.V., Ercole, E., ... & Groenewald, J.Z. (2018) Fungal Planet description sheets: 716–784. Persoonia 40: 240–393. https://doi.org/10.3767/persoonia.2018.40.10</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Currah, R., Tsuneda, A. & Murakami, S. (1993) Conidiogenesis in Oidiodendron periconioides and ultrastructure of ericoid mycorrhizas formed with Rhododendron brachycarpum. Canadian Journal of Botany 71: 1481–1485. https://doi.org/10.1139/b93-179</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dalpé, Y. (1986) Axenic synthesis of ericoid mycorrhiza in Vaccinium angustifolium Ait. by Oidiodendron species. New Phytologist 103: 391–396. https://doi.org/10.1111/j.1469-8137.1986.tb00624.x</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dalpé, Y. (1991) Statut endomycorhizien du genre Oidiodendron. Canadian Journal of Botany 69: 1712–1714. https://doi.org/10.1139/b91-217</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Darriba, D., Posada, D., Kozlov, A.M., Stamatakis, A., Morel, B. & Flouri, T. (2020) ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Molecular Biology and Evolution 37: 291–294. https://doi.org/10.1093/molbev/msz189</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Domsch, K.H., Grams, W. & Anderson<span lang="en-GB">,</span> T.H. (2007) Compendium of soil fungi second edition. IHW-Verlag, Eching.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Douglas, G., Heslin, M. & Reid<span lang="en-GB">,</span> C. (1989) Isolation of Oidiodendron maius from Rhododendron and ultrastructural characterization of synthesized mycorrhizas. Canadian Journal of Botany 67: 2206–2212. https://doi.org/10.1139/b89-280</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Edler, D., Klein, J., Antonelli, A. & Silvestro, D. (2021) raxmlGUI 2.0: A graphical interface and toolkit for phylogenetic analyses using RAxML. Methods in Ecology and Evolution 12: 373–377. http://dx.doi.org/10.1111/2041-210X.13512</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hambleton, S., Egger, K. & Currah, R. (1998) The genus Oidiodendron: species delimitation and phylogenetic relationships based on nuclear ribosomal DNA Analysis. Mycologia 90: 854–68. https://doi.org/10.2307/3761327</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hiradate, S. (2019) Properties of soils of the Ogasawara Islands: keys to understand past nature and find adequate management for future. Global Environmental Research 23: 29–36.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Johnston, P.R., Quijada, L., Smith, C.A., Baral, H.O., Hosoya, T., Baschien, C., Pärtel, K., Zhuang, W.Y., Haelewaters, D., Park, D., Carl, S., López-Giráldez, F., Wang, Z. & Townsend, J.P. (2019) A multigene phylogeny toward a new phylogenetic classification of Leotiomycetes. IMA Fungus 10: 1. https://doi.org/10.1186/s43008-019-0002-x</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. https://doi.org/10.1093/bib/bbx108</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kobae, Y. & Ohtomo, R. (2016) An improved method for bright-field imaging of arbuscular mycorrhizal fungi in plant roots. Soil Science and Plant Nutrition 62: 27–30. https://doi.org/10.1080/00380768.2015.1106923</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kozlov, A.M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. https://doi.org/10.1093/bioinformatics/btz305</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Lemoine, F., Domelevo Entfellner, J.B., Wilkinson, E., Correia, D., Dávila Felipe, M., De Oliveira, T. & Gascuel, O. (2018) Renewing Felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556: 452–456. https://doi.org/10.1038/s41586-018-0043-0</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Leopold, D.R. (2016) Ericoid fungal diversity: challenges and opportunities for mycorrhizal research. Fungal Ecology 24: 114–123. https://doi.org/10.1016/j. funeco.2016.07.004</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Morita, S., Kato, H., Iwasaki, N., Kusumoto, Y., Yoshida, K. & Hiradate, S. (2010) Unusually high levels of bio-available phosphate in the soils of Ogasawara Islands, Japan: Putative influence of seabirds. Geoderma 160: 155–164. https://doi.org/10.1016/j.geoderma.2010.09.008</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Murray, M.G. & Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research 8: 4321–4326. https://doi.org/10.1093/nar/8.19.4321</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">O’Donnell, K. & Cigelink, E. (1997) Two divergent intragenomic rRNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. https://doi.org/10.1006/mpev.1996.0376</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ogawa, Y., Suda, A., Kusama-Eguchi, K., Watanabe, K. & Tokumasu, S. (2005) Intraspecific groups of Umbelopsis ramanniana inferred from nucleotide sequences of nuclear rDNA internal transcribed spacer regions and sporangiospore morphology. Mycoscience 46: 343–351. https://dx.doi.org/10.1007/s10267-005-0257-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Raja, H., Schoch, C.L., Hustad, V., Shearer, C. & Miller, A. (2011) Testing the phylogenetic utility of MCM7 in the Ascomycota. MycoKeys 1: 63–94. https://doi.org/10.3897/mycokeys.1.1966</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rayner<span lang="en-GB">,</span> R. (1970) A mycological colour chart. Commonwealth Mycological Institute, Surrey & British Mycological Society, Kew.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rice, A. & Currah, R. (2005) Oidiodendron: A survey of the named species and related anamorphs of Myxotrichum. Studies in Mycology 53: 83–120. https://doi.org/10.3114/sim.53.1.83</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rice, A. & Currah, R. (2006) Oidiodendron maius: saprobe in Sphagnum peat, mutualist in ericaceous roots? In: Schulz, B.J.E., Boyle, C.J.C. & Sieber, T.N. (eds.) Microbial root endophytes. Springer Berlin / Heidelberg, Berlin, pp. 227–246. </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Robak, H. (1932) Investigations regarding fungi on Norwegian ground wood pulp and fungal infections at wood pulp mills. Saertrykk Nyt Mag. for Naturvidenskaberne 71: 185–330.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rodríguez-Andrade, E., Stchigel, A.M., Terrab, A., Guarro<span lang="en-GB">,</span> J. & Cano-Lira, J.F. (2019) Diversity of xerotolerant and xerophilic fungi in honey. IMA Fungus 10: 20. https://doi.org/10.1186/s43008-019-0021-7</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Sato, T., Uzuhashi, S., Hosoya, T. & Hosaka, K. (2010) A list of fungi found in the Bonin (Ogasawara) Islands. Ogasawara Research 35: 59–160.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Schmitt, I., Crespo, A., Divakar, P.K., Fankhauser, J.D., Herman-Sackett, E., Kalb, K., Nelsen, M.P., Nelson, N.A., Rivas-Plata, E., Shimp, A.D., Widhelm, T. & Lumbsch, H.T. (2009) New primers for promising single-copy genes in fungal phylogenetics and systematics. Persoonia 23: 35–40. https://doi.org/10.3767/003158509X470602</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Seifert, K.A., Hughes, S.J., Boulay, H. & Louis-Seize, G. (2007) Taxonomy, nomenclature and phylogeny of three cladosporium-like hyphomycetes, Sorocybe resinae, Seifertia azaleae and the Hormoconis anamorph of Amorphotheca resinae. Studies in Mycology 58: 235–245. https://doi.org/10.3114/sim.2007.58.09</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tian, W., Zhang, C.Q., Qiao, P. & Milne, R. (2011) Diversity of culturable ericoid mycorrhizal fungi of Rhododendron decorum in Yunnan, China. Mycologia 103: 703–709. https://doi.org/10.3852/10-296</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tokumasu, S. (1973) Notes on Japanese Oidiodendron. Transactions of the Mycological Society of Japan 14: 246–255</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tokumasu, S. (1996) Mycofloral succession on Pinus densiflora needles on a modern site. Mycoscience 37: 313–321. https://doi.org/10.1007/BF02461303</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Toyoda, T. (2003) Flora of Bonin Islands, 2nd Ed. Enlarged & Revised, Aboc & Co., Ltd., Kamakura, Japan. [in Japanese]</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wei, X., Chen, J., Zhang, C. & Pan, D. (2016) A new Oidiodendron maius strain isolated from Rhododendron fortunei and its effects on nitrogen uptake and plant growth. Frontiers in Microbiology 7: 1327. https://doi.org/10.3389/fmicb.2016.01327</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">White, T.J., Bruns, T., Lee, S. & Taylor, J.W. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, Y.J. (eds.) PCR Protocols: A Guide to Methods and Application. Academic Press, San Diego, pp 315–322.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Xiao, G. & Berch, S. (1995) The ability of known ericoid mycorrhizal fungi to form mycorrhizae with Gaultheria shallon. Mycologia 87: 467–70. https://doi.org/10.2307/3760763</span></span></span>