Abstract
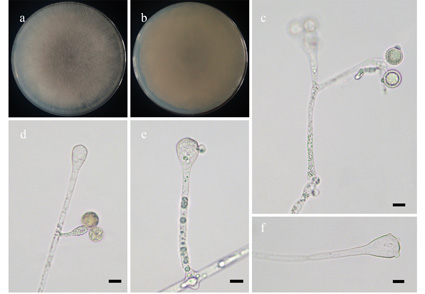
Species of Cunninghamella are ubiquitous in soil, flower and plant debris. Its metabolites have various biological activities, such as antifungal and antibacterial functions. In this study, we describe a new species C. verrucosa from soil in Guangdong Province, China. This species is characterized by one to several broken pedicels on the surface of clavate vesicles, unbranched or simply branched sporangiophores, and globose sporangiola. Phylogenetic analyses strongly show that C. verrucosa is sister to C. clavata. Together with this new species, a total of 22 taxa, including 19 species and three varieties, has been described in Cunninghamella. Accordingly, an updated key to the taxa of Cunninghamella is also provided in this study.
References
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Alves, A., de Souza, C., de Oliveira, R., Cordeiro, T. & de Santiago, A. (2017) Cunninghamella clavata from Brazil: a new record for the western hemisphere. Mycotaxon 132: 381–389. https://doi.org/10.5248/132.381</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Cha, C.J., Doerge, D.R. & Cerniglia, C.E. (2001) Biotransformation of malachite green by the fungus Cunninghamella elegans. Applied and Environmental Microbiololgy 67: 4358–4360. https://doi.org/10.1128/aem.67.9.4358-4360.2001</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dai, Y.C. (2010) Hymenochaetaceae (Basidiomycota) in China. Fungal Diversity 45: 131–343. https://doi.org/10.1007/s13225-010-0066-9</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Elkhateeb, W.A., Elnahas, M.O. & Daba, G.M. (2021) Bioactive metabolites of Cunninghamella, biodiversity to biotechnology. Journal of Pharmaceutics and Pharmacology Research 4: 1–5. https://doi.org/10.31579/2693-7247/036</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">GBIF-Secretariat (2021) GBIF Backbone Taxonomy. Checklist dataset. Via GBIF.org. Available at https://doi.org/10.15468/39omei. (Accessed on April 25, 2022)</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Guo, J., Wang, H., Liu, D., Zhang, J.N., Zhao, Y.H., Liu, T.X. & Xin, Z.H. (2015) Isolation of Cunninghamella bigelovii sp. nov. CGMCC 8094 as a new endophytic oleaginous fungus from Salicornia bigelovii. Mycological Progress 14: 1–8. https://doi.org/10.1007/s11557-015-1029-z</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hallur, V., Prakash, H., Sable, M., Preetam, C., Purushotham, P., Senapati, R., Shankarnarayan, S.A., Bag, N.D. & Rudramurthy, S.M. (2021) Cunninghamella arunalokei a new species of Cunninghamella from India causing disease in an immunocompetent individual. Journal of Fungi 7: 670. https://doi.org/10.3390/jof7080670</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hyde, K.D., Hongsanan, S., Jeewon, R., Bhat, J.D., McKenzie, E.H.C., Jones, E.B.G., Phookamsak, R., Ariyawansa, H.A., Boonmee, S., Zhao, Q., Abdel-Aziz, F.A., Abdel-Wahab, M.A., Banmai, S., Chomnunti, P., Cui, B.K., Daranagama, D.A., Das, K., Dayarathne, M.C., De Silva, N.I., Dissanayake, A.J., Doilom, M., Ekanayaka, A.H., Gibertoni, T.B., Go´es-Neto, A., Huang, S.K., Jayasiri, S.C., Jayawardena, R., Konta, S., Lee, H.B., Li, W.J., Lin, C.G., Liu, J.K., Lu, Y.Z., Luo, Z.L., Manawasinghe, I.S., Manimohan, P., Mapook, A., Niskanen, T., Norphanphoun, C., Papizadeh, M., Perera, R.H., Phukhamsakda, C., Richter, C., de A. Santiago, A.L.C.M., Drechsler-Santos, E.R., Senanayake, I.C. & Tanaka, K. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. https://doi.org/10.1007/s13225-016-0373-x</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Kearse, M,, Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S. & Duran, C. (2012) Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. https://doi.org/10.1093/bioinformatics/bts199</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, C.W., Liou, G.Y. & Chien, C.Y. (2005) New records of the genus Cunninghamella (Mucorales) in Taiwan. Fungal Science 20: 1–9</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, X.Y., Huang, H. & Zheng, R.Y. (2001) Relationships within Cunninghamella based on sequence analysis of ITS rDNA. Mycotaxon 80: 77–95</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Nguyen, T.T.T, Choi, Y.J. & Lee, H.B. (2017) Isolation and characterization of three unrecorded Zygomycete fungi in Korea: Cunninghamella bertholletiae, Cunninghamella echinulata, and Cunninghamella elegans. Mycobiology 45: 318–326. https://doi.org/10.5941/MYCO.2017.45.4.318</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Nie, Y., Cai, Y., Gao, Y., Yu, D.S., Wang, Z.M., Liu, X.Y. & Huang, B. (2020a) Three new species of Conidiobolus sensu stricto from plant debris in eastern China. MycoKeys 73: 133–149. https://doi.org/10.3897/mycokeys.73.56905</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Nie, Y., Yu, D.S., Wang, C.F., Liu, X.Y. & Huang, B. (2020b) A taxonomic revision of the genus Conidiobolus (Ancylistaceae, Entomophthorales): four clades including three new genera. MycoKeys 66: 55–81. https://doi.org/10.3897/mycokeys.66.46575</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Posada, D. & Crandall, K.A. (1998) ModelTest: testing the model of DNA substitution. Bioinformatics 14: 817–818. https://doi.org/10.1093/bioinformatics/14.9.817</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Spatafora, J.W., Chang, Y., Benny, G.L., Lazarus, K., Smith, M.E., Berbee, M.L., Bonito, G., Corradi, N., Grigoriev, I. & Gryganskyi, A. (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108: 1028–1046. https://doi.org/10.3852/16-042</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Suwannarach, N., Kumla, J., Supo, C., Honda, Y., Nakazawa, T., Khanongnuch, C. & Wongputtisin, P. (2021) Cunninghamella saisamornae (Cunninghamellaceae, Mucorales), a new soil fungus from northern Thailand. Phytotaxa 509: 291–300. https://doi.org/10.11646/phytotaxa.509.3.4</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Swofford, D.L. (2002) PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods); Version 4.0b10; Sinauer Associates: Sunderland, MA, USA.</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Voigt, K., James, T.Y., Kirk, P.M., da Santiago, A.L.C.M., Waldman, B., Griffith, G.W., Fu, M., Radek, R., Strassert, J.F.H., Wurzbacher, C., Jerônimo, G.H., Simmons, D.R., Seto, K., Gentekaki, E., Hurdeal, V.G., Hyde, K.D., Nguyen, T.T.T. & Lee, H.B. (2021) Early-diverging fungal phyla: taxonomy, species concept, ecology, distribution, anthropogenic impact, and novel phylogenetic proposals. Fungal Diversity 109: 59–98. https://doi.org/10.1007/s13225-021-00480-y</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Walther, G., Paw?owska, J., Alastruey-Izquierdo, A., Wrzosek, M., Rodriguez-Tudela, J.L., Dolatabadi, S., Chakrabarti, A. & de Hoog, G.S. (2013) DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia 30: 11–47. https://doi.org/10.3767/003158513X665070</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Walther, G., Wagner, L. & Kurzai, O. (2019) Updates on the taxonomy of Mucorales with an emphasis on clinically important taxa. Journal of Fungi 5: 106. https://doi.org/10.3390/jof5040106</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wang, K., Dai, Y.C., Jia, Z.F., Li, T.H., Liu, T.Z., Dorji, P., Reyim, M., Sun, G.Y., Tolgor, B., Wei, S.L., Yang, Z.L., Yuan, H.S., Zhang, X.G. & Cai, L. (2021) Overview of China’s nomenclature novelties of fungi in the new century (2000-2020). Mycosystema 40: 822–833. https://doi.org/10.13346/j.mycosystema.210064</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wang, K., Liu, F. & Cai, L. (2022) A name list of common agricultural phytopathogenic fungi in China. Mycosystema 41: 361–386. https://doi.org/10.13346/j.mycosystema.210483</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenies. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR Protocols: a guide to methods and applications. Academic Press, Inc, San Diego, California, pp. 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wijayawardene, N., Hyde, K., Al-Ani, L., Tedersoo, L., Haelewaters, D., Rajeshkumar, K., Zhao, R., Aptroot, A., Leontyev, D. & Saxena, R. (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11: 1060–1456. https://doi.org/10.5943/mycosphere/11/1/8</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wu, F., Zhou, L.W., Josef, V. & Dai, Y.C. (2022) Global diversity and systematics of Hymenochaetaceae with poroid hymenophore. Fungal Diversity 113: 1–192. https://doi.org/10.1007/s13225-021-00496-4</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Yu, J., Walther, G., van Diepeningen, A.H.G, Gerrits, V.D.E., Li, R.Y,, Moussa, T., Almaghrabi, O. & De Hoog, G.S. (2014) DNA barcoding of clinically relevant Cunninghamella species. Medical Mycology 53: 99–106. https://doi.org/10.1093/mmy/myu079</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, Z.Y., Zhao, Y.X., Shen, X., Chen, W.H., Han, Y.F., Huang, J.Z. & Liang, Z.Q. (2020) Molecular phylogeny and morphology of Cunninghamella guizhouensis sp. nov. (Cunninghamellaceae, Mucorales), from soil in Guizhou, China. Phytotaxa 455: 31–39. https://doi.org/10.11646/phytotaxa.455.1.4</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhao, H., Lv, M.L., Liu, Z., Zhang, M.Z., Wang, Y.N., Ju, X., Song, Z., Ren, L.Y., Jia, B.S., Qiao, M. & Liu, X.Y. (2021a) High-yield oleaginous fungi and high-value microbial lipid resources from Mucoromycota. BioEnergy Research 14: 1196–1206. https://doi.org/10.1007/s12155-020-10219-3</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhao, H., Nie, Y., Zong, T.K., Dai, Y.C. & Liu, X.Y. (2022) Three new species of Absidia (Mucoromycota) from China based on phylogeny, morphology and physiology. Diversity 14: 132. https://doi.org/10.3390/d14020132</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhao, H., Zhu, J., Zong, T.K., Liu, X.L., Ren, L.Y., Lin, Q., Qiao, M., Nie, Y., Zhang, Z.D. & Liu, X.Y. (2021b) Two new species in the family Cunninghamellaceae from China. Mycobiology 49: 142–150. https:// doi.org/10.1080/12298093.2021.1904555</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zheng, R.Y. & Chen, G.Q. (1998) Cunninghamella clavata sp. nov., a fungus with an unusual type of branching of sporophore. Mycotaxon 69: 187–198</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zheng, R.Y. & Chen, G.Q (2001) A monograph of Cunninghamella. Mycotaxon 80: 1–75</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zheng, R.Y., Chen, G.Q., Huang, H. & Liu, X.Y. (2007) A monograph of Rhizopus. Sydowia 59: 273–372</span></span></span>
<span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zong, T.K., Zhao, H., Liu, X.L., Ren, L.Y., Zhao, C.L. & Liu, X.Y. (2021) Taxonomy and phylogeny of four new species in Absidia (Cunninghamellaceae, Mucorales) from China. Frontiers in Microbiology 12: 2181. https:// doi.org/10.3389/fmicb.2021.677836</span></span>