Abstract
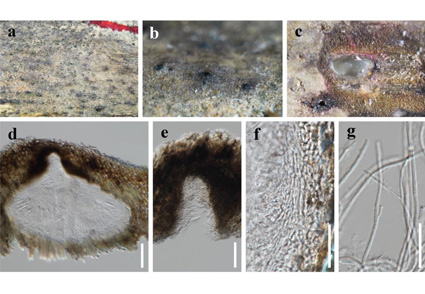
During an investigation of ascomycetous fungi on bamboos in Sichuan province, China, a monotypic genus, Pseudokeissleriella, collected from dead culms of bamboos is introduced to accommodate P. bambusicola. Pseudokeissleriella bambusicola is characterized by having subglobose to globose, glabrous ascomata, and hyaline, septate, fusiform ascospores with subobtuse ends and a swollen upper cell, surrounded by a mucilaginous sheath with center depression. The phylogenetic analyses based on multi-gene matrix of SSU, ITS, LSU, tef-1α sequences showed that P. bambusicola presented a distinct lineage sister to Katumotoa and Neoophiosphaerella in Lentitheciaceae. The establishment of new taxa were justified by morphological and phylogenetic evidences. Morpho-phylogenetic differences between Pseudokeissleriella and some related genera Katumotoa, Keissleriella, and Neoophiosphaerella are discussed. Descriptions, illustrations, and notes for the new taxa are provided.
References
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Calabon, M.S., Jones, E.B.G., Hyde, K.D., Boonmee, S., Tibell, S., Tibell, L., Pang, K.L. & Phookamsak, R. (2021) Phylogenetic assessment and taxonomic revision of Halobyssothecium and Lentithecium (Lentitheciaceae, Pleosporales). Mycological Progress 20: 701–720. https://doi.org/10.1007/s13225-014-0278-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Chomnunti, P., Hongsanan, S., Aguirre, Hudson, B., Tian, Q., Persoh, D., Dhami, M.K., Alias, A.S., Xu, J.C., Liu, X.Z., Stadler, M. & Hyde, K.D. (2014) The sooty moulds. Fungal Diversity 66: 1–36. https://doi.org/10.1007/s13225-014-0278-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Crous, P.W., Shivas, R.G., Quaedvlieg, W., van der Bank, M., Zhang, Y., Summerell, B.A., Guarro, J., Wingfield, M.J., Wood, A.R., Alfenas, A.C., Braun, U., Cano-Lira, J.F., García, D., Marin-Felix, Y., Alvarado, P., Andrade, J.P., Armengol, J., Assefa, A., den Breeÿen, A., Camele, I., Cheewangkoon, R., De Souza, J.T., Duong, T.A., Esteve-Raventós, F., Fournier, J., Frisullo, S., García-Jiménez, J., Gardiennet, A., Gené, J., Hernández-Restrepo, M., Hirooka, Y., Hospenthal, D.R., King, A., Lechat, C., Lombard, L., Mang, S.M., Marbach, P.A.S., Marincowitz, S., Marin-Felix, Y., Montaño-Mata, N.J., Moreno, G., Perez, C.A., Pérez Sierra, A.M., Robertson, J.L., Roux, J., Rubio, E., Schumacher, R.K., Stchigel, A.M., Sutton, D.A., Tan, Y.P., Thompson, E.H., van der Linde, E., Walker, A.K., Walker, D.M., Wickes, B.L., Wong, P.T.W. & Groenewald, J.Z. (2014) Fungal Planet Description Sheets: 214–280. Persoonia 32: 184–306. https://doi.org/10.3767/003158514X682395</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dai, D.Q., Tang, L.Z., Wang, H.J.B.C. & Prospects, F. (2018) A review of Bambusicolous ascomycetes. Bamboo - Current and Future Prospects 10: 165–183. https://doi.org/10.5772/intechopen.76463</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dayarathne, M.C., Wanasingh, D.N., Jones, E.B.G., Chomnunti, P. & Hyde, K.D. (2018) A novel marine genus, Halobyssothecium (Lentitheciaceae) and epitypification of Halobyssothecium obiones comb. nov. Mycological Progress 17: 1161–1171. https://doi.org/10.1007/s11557-018-1432-3</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Devadatha, B., Calabon, M.S., Abeywickrama, P.D., Hyde, K.D. & Jones, E.B.G. (2019) Molecular data reveals a new holomorphic marine fungus, Halobyssothecium estuariae, and the asexual morph of Keissleriella phragmiticola. Mycology 11: 167–183. https://doi.org/10.1080/21501203.2019.1700025</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Dissanayake, A.J., Bhunjun, C.S., Maharachchikumbura, S.S.M. & Liu, J.K. (2020) Applied aspects of methods to infer phylogenetic relationships amongst fungi. Mycosphere 11: 2652–2676. https://doi.org/10.5943/mycosphere/11/1/18</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hall, T.A. (1999) BioEdit: A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. 41: 95–98. https://doi.org/10.1021/bk-1999-0734.ch008</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hirayama, K., Tanaka, K., Raja, H.A., Miller, A.N. & Shearer, C.A. (2010) A molecular phylogenetic assessment of Massarina ingoldiana sensu lato. Mycologia 102: 729–746. https://doi.org/10.3852/09-230</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hongsanan, S., Hyde, K.D., Phookamsak, R., Wanasinghe, D.N., McKenzie, E.H.C., Sarma, V.V., Boonmee, S., Lucking, R., Bhat, D.J., Liu, N.G., Tennakoon, D.S., Pem, D.S., Karunarathna, A., Jiang, S.H., Jones, E.B.G., Phillips, A.J.L., Manawasinghe, I.S., Tibpromma, S., Jayasiri, S.C., Sandamali, D.S., Jayawardena, R.S., Wijayawardene, N.N., Ekanayaka, A.H., Jeewon, R., Lu, Y.Z., Dissanayake, A.J., Zeng, X.Y., Luo, Z.L., Tian, Q., Phukhamsakda, C., Thambugala, K.M., Dai, D.Q., Chethana, K.W.T., Samarakoon, M.C., Ertz, D., Bao, D.F., Doilom, M., Liu, J.K., Perez-Ortega, S., Suija, A., Senwanna, C., Wijesinghe, S.N., Konta, S., Niranjan, M., Zhang, S.N., Ariyawansa, H.A., Jiang, H.B., Zhang, J.F., Norphanphoun, C., de Silva, N.I., Thiyagaraja, V., Zhang, H., Bezerra, J.D.P., Miranda-Gonzalez, R., Aptroot, A., Kashiwadani, H., Harishchandra, D., Serusiaux, E., Aluthmuhandiram, J.V.S., Abeywickrama, P.D., Devadatha, B., Wu, H.X., Moon, K.H., Gueidan, C., Schumm, F., Bundhun, D., Mapook, A., Monkai, J., Chomnunti, P., Suetrong, S., Chaiwan, N., Dayarathne, M.C., Yang, J., Rathnayaka, A.R., Bhunjun, C.S., Xu, J.C., Zheng, J.S., Liu, G.L., Feng, Y. & Xie, N. (2020) Refined families of Dothideomycetes: Dothideomycetidae and Pleosporomycetidae. Mycosphere 11: 1553–2107. https://doi.org/10.5943/mycosphere/11/1/13</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Hyde, K.D., Dong, Y., Phookamsak, R., Jeewon, R., Bhat, D.J., Jones, E.B.G., Liu, N.G., Abeywickrama, P.D., Mapook, A., Wei, D., Perera, R.H., Manawasinghe, I.S., Pem, D., Bundhun, D., Karunarathna, A., Ekanayaka ,A.H., Bao, D.F., Li, J., Samarakoon, M.C., Chaiwan, N., Lin, C.G., Phutthacharoen, K., Zhang, S.N., Senanayake, I.C., Goonasekara, I.D., Thambugala, K.M., Phukhamsakda, C., Tennakoon, D.S., Jiang, H.B., Yang, J., Zeng, M., Huanraluek, N., Liu, J.K., Wijesinghe, S.N., Tian, Q., Tibpromma, S., Brahmanage, R.S., Boonmee, S., Huang, S.K., Thiyagaraja, V., Lu, Y.Z., Jayawardena, R.S., Dong, W., Yang, E.F., Singh, S.K., Singh, S.M., Rana, S., Lad, S.S., Anand, G., Devadatha, B., Niranjan, M., Sarma, V.V., Liimatainen, K., Aguirre-Hudson, B., Niskanen, T., Overall, A., Alvarenga, R.L.M., Gibertoni, T.B., Pfliegler, W.P., Horváth, E., Imre, A., Alves, A.L., da Silva Santos, A.C., Tiago, P.V., Bulgakov, T.S., Wanasinghe, D.N., Bahkali, A.H., Doilom, M., Elgorban, A.M., Maharachchikumbura, S.S.N., Rajeshkumar, K.C., Haelewaters, D., Mortimer<span lang="en-GB">,</span> P.E., Zhao, Q., Lumyong, S., Xu, J. & Sheng, J. (2020) Fungal diversity notes 1151–1276: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Diversity 100: 5–277. https://doi.org/10.1007/s13225-020-00439-5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Katoh, K. & Standley, D.M. (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution 30: 772–780. https://doi.org/10.1093/molbev/mst010</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Knapp, D.G., Kovacs, G.M., Zajta, E., Groenewald, J.Z. & Crous, P.W. (2015) Dark septate endophytic pleosporalean genera from semiarid areas. Persoonia 35: 87–100. https://doi.org/10.3767/003158515X687669</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Jeewon, R. & Hyde, K.D. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. https://doi.org/10.5943/mycosphere/7/11/4</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Jiang, H.B., Phookamsak, R., Doilom, M., Mortimer, P.E., Xu, J.C., Lumyong, S., Hyde, K.D. & Karunarathna, S.C. (2019) Taxonomic and phylogenetic characterizations of Keissleriella bambusicola sp. nov. (Lentitheciaceae, Pleosporales) from Yunnan, China. Phytotaxa 423: 129–144. https://doi.org/10.11646/phytotaxa.423.3.2</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Lanfear, R., Frandsen, P.B., Wright, A.M., Senfeld, T. & Calcott, B. (2017) PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Molecular Biology and Evolution 34: 772–773. https://doi.org/10.1093/molbev/msw260</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Li, G.J., Hyde, K.D., Zhao, R.L., Hongsanan, S., Abdel Aziz, F.A., Abdel-Wahab, M.A., Alvarado, P., Alves Silva, G., Ammirati, J.F., Ariyawansa, H.A., Baghela, A., Bahkali, A.H., Beug, M., Bhat, D.J., Bojantchev, D., Boonpratuang, T., Bulgakov, T.S., Camporesi, E., Boro, M.C., Ceska, O., Chakraborty, D., Chen, J.J., Chethana, K.W.T., Chomnunti, P., Consiglio, G., Cui, B.K., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Das, K., Dayarathne, M.C., De Crop, E., De Oliveira, R.J.V., de Souza, C.A.F., de Souza, J.I., Dentinger, B.T.M., Dissanayake, A.J., Doilom, M., Drechsler-Santos, E.R., Ghobad-Nejhad, M., Gilmore, S.P., Góes-Neto, A., Gorczak, M., Haitjema, C.H., Hapuarachchi, K.K., Hashimoto, A., He, M.Q., Henske, J.K., Hirayama, K., Iribarren, M.J., Jayasiri, S.C., Jayawardena, R.S., Jeon, S.J., Jerônimo, G.H., Jesus, A.L., Jones, E.B.G., Kang, J.C., Karunarathna, S.C., Kirk, P.M., Konta, S., Kuhnert, E., Langer, E., Lee, H.S., Lee, H.B., Li, W.J., Li, X.H., Liimatainen, K., Lima, D.X., Lin, C.G., Liu, J.K., Liu, X.Z., Liu, Z.Y., Luangsa-ard, J.J., Lücking, R., Lumbsch, H.T., Lumyong, S., Leaño, E.M., Marano, A.V., Matsumura, M., McKenzie, E.H.C., Mongkolsamrit, S., Mortimer, P.E., Nguyen, T.T.T., Niskanen, T., Norphanphoun, C., O’Malley, M.A., Parnmen, S., Paw?owska, J., Perera, R.H., Phookamsak, R., Phukhamsakda, C., Pires-Zottarelli, C.L.A., Raspé, O., Reck, M.A., Rocha, S.C.O., de Santiago, A.L.C.M.A., Senanayake, I.C., Setti, L., Shang, Q.J., Singh, S.K., Sir, E.B., Solomon, K.V., Song, J., Srikitikulchai, P., Stadler, M., Suetrong, S., Takahashi, H., Takahashi, T., Tanaka, K., Tang, L.P., Thambugala, K.M., Thanakitpipattana, D., Theodorou, M.K., Thongbai, B., Thummarukcharoen, T., Tian, Q., Tibpromma, S., Verbeken, A., Vizzini, A., Vlasák, J., Voigt, K., Wanasinghe, D.N., Wang, Y., Weerakoon, G., Wen, H.A., Wen, T.C., Wijayawardene, N.N., Wongkanoun, S., Wrzosek, M., Xiao, Y.P., Xu, J.C., Yan, J.Y., Yang, J., Da Yang, S., Hu, Y., Zhang, J.F., Zhao, J., Zhou, L.W., Peršoh, D., Phillips, A.J.L. & Maharachchikumbura, S.S.N. (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 78: 1–237. https://doi.org/10.1007/s13225-016-0366-9</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, J.K., Chomnunti, P., Cai, L., Phookamsak, R., Chukeatirote, E., Jones, E.B.G., Moslem, M. & Hyde, K.D. (2010) Phylogeny and morphology of Neodeightonia palmicola sp. nov. from palms. Sydowia 62: 261–276</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, J.K., Hyde, K.D., Jones, E.B.G., Ariyawansa, H.A., Bhat, D.J., Boonmee, S., Maharachchikumbura, S.S.N., McKenzie, E.H.C., Phookamsak, R., Phukhamsakda, C., Shenoy, B.D., Abdel-Wahab, M.A., Buyck, B., Chen, J., Thilini Chethana, K.W., Singtripop, C., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Dissanayake, A.J., Doilom, M., D’souza, M.J., Fan, X.L., Goonasekara, I.D., Hirayama, K., Hongsanan, S., Jayasiri, S.C., Jayawardena, R.S., Karunarathna, S.C., Li, W.J., Mapook, A., Norphanphoun, C., Pang, K.L., Perera, R.H., Peršoh, D., Pinruan, U., Senanayake, I.C., Somrithipol, S., Suetrong, S., Tanaka, K., Thambugala, K.M., Tian, Q., Tibpromma, S., Udayanga, D., Wijayawardene, N.N., Wanasinghe, D., Wisitrassameewong, K., Zeng, X.Y., Abdel-Aziz, F.A., Adam?ík, S., Boonyuen, A.H.B., Bulgakov, T., Callac, P., Chomnunti, P., Greiner, K., Hashimoto, A., Hofstetter, V., Kang, J.C., Lewis, D., Li, X.H., Liu, X.Z., Liu, Z.Y., Matsumura, M., Mortimer, P.E., Rambold, G., Randrianjohany, E., Sato, G., Sri-Indrasutdhi, V., Cheng, M.T., Verbeken, A., Brackel, W.V., Wang, Y., Wen, T.C., Xu, J.C., Yan, J.Y., Zhao, R.L. & Camporesi, E. (2015) Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. https://doi.org/10.1007/s13225-015-0324-y</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Liu, Z.P., Zhang, S.N., Cheewangkoon, R., Zhao, Q. & Liu, J.K. (2022) Crassoascoma gen. nov. (Lentitheciaceae, Pleosporales): Unrevealing Microfungi from the Qinghai-Tibet plateau in China. Diversity 14: 15. https://doi.org/10.3390/d14010015</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Luo, Z.L., Bahkali, A.H., Liu, X.Y., Phookamsak, R., Zhao, Y.C., Zhou, D.Q., Su, H.Y. & Hyde, K.D. (2016) Poaceascoma aquaticum sp nov (Lentitheciaceae), a new species from submerged bamboo in freshwater. Phytotaxa 253: 71–80. https://doi.org/10.11646/phytotaxa.253.1.5</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Maddison, W.P. & Maddison, D.R. (2014) Mesquite: a modular system for evolutionary analysis. Version 3.01. Available from: http://mesquiteproject.org (accessed 15 April 2022). </span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES science gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010. New Orleans, pp. 1–8. https://doi.org/10.1109/GCE.2010.5676129</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Phookamsak, R., Manamgoda, D.S., Li, W.J., Dai, D.Q., Singtripop, C. & Hyde, K.D. (2015) Poaceascoma helicoides gen et sp. nov., a new genus with scolecospores in Lentitheciaceae. Cryptogamie Mycologie 36: 225–236. https://doi.org/10.7872/crym/v36.iss2.2015.225</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Phookamsak, R., Hyde, K.D., Jeewon, R., Bhat, D.J., Jones, E.B.G., Maharachchikumbura, S.S.N., Raspé, O., Karunarathna, S.C., Wanasinghe, D.N., Hongsanan, S., Doilom, M., Tennakoon, D.S., Machado, A.R., Firmino, A.L., Ghosh, A., Karunarathna, A., Meši?, A., Dutta, A.K., Thongbai, B., Devadatha, B., Norphanphoun, C., Senwanna, C., Wei, D., Pem, D., Ackah, F.K., Wang, G.N., Jiang, H.B., Madrid, H., Lee, H.B., Goonasekara, I.D., Manawasinghe, I.S., Kušan, I., Cano, J., Gené, J., Li, J., Das, K., Acharya, K., Raj, K.N.A., Latha, K.P.D., Chethana, K.W.T., He, M.Q., Due,M., Jadan, M., Martín, M.P., Samarakoon, M.C., Dayarathne, M.C., Raza, M., Park, M.S., Telleria, M.T., Chaiwan, N., Mato?ec, N., de Silva, N.I., Pereira, O.L., Singh, P.N., Manimohan, P., Uniyal, P., Shang, Q.J., Bhatt, R.P., Perera, R.H., Alvarenga, R.L.M., Nogal-Prata, S., Singh, S.K., Vadthanarat, S., Oh, S.Y., Huang, S.K., Rana, S., Konta, S., Paloi, S., Jayasiri, S.C., Jeon, S.J., Mehmood, T., Gibertoni, T.B., Nguyen, T.T.T., Singh, U., Thiyagaraja, V., Sarma, V.V., Dong, W., Yu, X.D., Lu, Y.Z., Lim, Y.W., Chen, Y., Tkal?ec, Z., Zhang, Z.F., Luo, Z.L., Daranagama, D.A., Thambugala, K.M., Tibpromma, S., Camporesi, E., Bulgakov, T.S., Dissanayake, A.J., Senanayake, I.C., Dai, D.Q., Tang, L.Z., Khan, S., Zhang, H., Promputtha, I., Cai, L., Chomnunti, P., Zhao, R.L., Lumyong, S., Boonmee, S., Wen, T.C., Mortimer, P.E. & Xu, J.C. (2019) Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Diversity 95: 1–273. https://doi.org/10.1007/s13225-019-00421-w</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Quaedvlieg, W., Verkley, G.J., Shin, H.D., Barreto, R.W., Alfenas, A.C., Swart, W.J., Groenewald, J.Z. & Crous, P.W. (2013) Sizing up Septoria. Studies in Mycology 75: 307–390. https://doi.org/10.3114/sim0017</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rathnayaka, A.R. Dayarathne, M.C., Maharachchikumbura, S.S.N., Liu, J.K., Tennakoon, D.S. & Hyde, K.D. (2019) Introducing Seriascoma yunnanense sp. nov. (Occultibambusaceae, Pleosporales) based on evidence from morphology and phylogeny. Asian Journal of Mycology 2: 245–253. https://doi.org/10.5943/ajom/2/1/15</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Rehner, S.A. & Buckley, E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-alpha sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. https://doi.org/10.3852/mycologia.97.1.84</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D.L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Senanayake, I.C., Rathnayaka, A.R., Marasinghe, D.S., Calabon, M.S., Gentekaki, E., Lee, H.B., Hurdeal, V.G., Pem, D., Dissanayake, L.S., Wijesinghe, S.N., Bundhun, D., Nguyen, T.T., Goonasekara, I.D., Abeywickrama, P.D., Bhunjun, C.S., Jayawardena, R.S., Wanasinghe, D.N., Jeewon, R., Bhat, D.J. & Xiang, M.M. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Singtripop, C., Camporesi, E., Ariyawansa, H.A., Wanasinghe, D.N., Bahkall, A.H., Chomnunti, P., Boonmee, S. & Hyde, K.D. (2015) Keissleriella dactylidis, sp. nov., from Dactylis glomerata and its phylogenetic placement. Scienceasia 41: 295–304. https://doi.org/10.2306/scienceasia1513-1874.2015.41.295</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Stamatakis, A., Hoover, P. & Rougemont, J. (2008) A rapid bootstrap algorithm for the RAxML Web servers. Studies in Mycology 57: 758–771. https://doi.org/10.1080/10635150802429642</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Su, H.Y., Luo, Z.L., Liu, X.Y., Su, X.J., Hu, D.M., Zhou ,D.Q., Bahkali, A.H. & Hyde, K.D. (2016) Lentithecium cangshanense sp. nov. (Lentitheciaceae) from freshwater habitats in Yunnan Province, China. Phytotaxa 267: 61–69. https://doi.org/10.11646/phytotaxa.267.1.6</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Suetrong, S., Schoch, C.L., Spatafora, J.W., Kohlmeyer, J., Volkmann Kohlmeyer, B., Sakayaroj, J., Phongpaichit, S., Tanaka, K., Hirayama, K. & Jones, E.B.G. (2009) Molecular systematics of the marine Dothideomycetes. Studies in mycology 64: 155–173. https://doi.org/10.3114/sim.2009.64.09</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Sun, Y., Goonasekara, I.D., Thambugala, K.M., Jayawardena, R.S., Wang, Y. & Hyde, K.D. (2020) Distoseptispora bambusae sp. nov. (Distoseptisporaceae) on bamboo from China and Thailand. Biodiversity Data Journal 8: e53678. https://doi.org/10.3897/BDJ.8.e53678</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Swofford, D. (2002) PAUP*. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4.0b10, vol Version 4.0. https://doi.org/10.1111/j.0014-3820.2002.tb00191.x</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tanaka, K. & Harada, Y. (2005) Bambusicolous fungi in Japan (6): Katumotoa, a new genus of phaeosphaeriaceous ascomycetes. Mycoscience 46: 313–318. https://doi.org/10.1007/S10267-005-0251-Y</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tanaka, K., Hirayama, K., Yonezawa, H., Sato, G., Toriyabe, A., Kudo, H., Hashimoto, A., Matsumura, M., Harada, Y., Kurihara, Y., Shirouzu, T. & Hosoya, T. (2015) Revision of the Massarineae (Pleosporales, Dothideomycetes). Studies in Mycology 82: 75–136. https://doi.org/10.1016/j.simyco.2015.10.002</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Tibpromma, S., Hyde, K.D., Jeewon, R., Maharachchikumbura, S.S.N., Liu, J.K., Bhat, D.J., Jones, E.B.G., McKenzie, E.H.C., Camporesi, E., Bulgakov, T.S., Doilom, M., de Azevedo Santiago, A.L.C.M., Das, K., Manimohan, P., Gibertoni, T.B., Lim, Y.W., Ekanayaka, A.H., Thongbai, B., Lee, H.B., Yang, J.B., Kirk, P.M., Sysouphanthong, P., Singh, S.K., Boonmee, S., Dong, W., Raj, K.N.A., Latha, K.P.D., Phookamsak, R., Phukhamsakda, C., Konta, S., Jayasiri, S.C., Norphanphoun, C., Tennakoon, D.S., Li, J.F., Dayarathne, M.C., Perera, R.H., Xiao, Y.P., Wanasinghe, D.N., Senanayake, I.C., Goonasekara, I.D., de Silva, N.I., Mapook, A., Jayawardena, R.S., Dissanayake, A.J., Manawasinghe, I.S., Chethana, K.W.T., Luo, Z.L., Hapuarachchi, K.K., Baghela, A., Soares, A.M., Vizzini, A., Meiras-Ottoni, A., Meši?, A., Dutta, A.K., de Souza, C.A.F., Richter, C., Lin, C.G., Chakrabarty, D., Daranagama, D.A. & Lima, D.X. (2017) Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 83: 1–261. https://doi.org/10.1007/s13225-017-0378-0</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Toju, H., Tanabe, A.S., Yamamoto, S. & Sato, H. (2012) High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One 7: e40863. https://doi.org/10.1371/journal.pone.0040863</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wanasinghe, D.N., Jones, E.B.G., Camporesi, E., Boonmee, S., Ariyawansa, H.A., Wijayawardene, N.N., Mortimer, P.E., Xu, J., Yang, J.B. & Hyde, K.D. (2014) An exciting novel member of Lentitheciaceae in Italy from Clematis vitalba. Cryptogamie Mycologie 35: 323–337. https://doi.org/10.7872/crym.v35.iss4.2014.323</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wanasinghe, D.N., Phukhamsakda, C., Hyde, K.D., Jeewon, R., Lee, H.B., Jones, E.B.G., Tibpromma, S., Tennakoon, D.S., Dissanayake, A.J., Jayasiri, S.C., Gafforov, Y., Camporesi, E., Bulgakov, T.S., Ekanayake, A.H., Perera, R.H., Samarakoon, M.C., Goonasekara, I.D., Mapook, A., Li, W.J., Senanayake, I.C., Li, J., Norphanphoun, C., Doilom, M., Bahkali, A.H., Xu, J., Mortimer, P.E., Tibell, L., Tibell, S. & Karunarathna, S.C. (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89: 1–236. https://doi.org/10.1007/s13225-018-0395-7</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal Rna genes for phylogenetics. In: PCR Protocols. pp 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Wijayawardene, N.N., Hyde, K.D., Bhat, D.J., Goonasekara, I.D., Nadeeshan, D., Camporesi, E., Schumacher, R.K. & Wang, Y. (2015) Additions to brown spored coelomycetous taxa in Massarinae, Pleosporales: introducing Phragmocamarosporium gen. nov and Suttonomyces gen. nov. Cryptogamie Mycologie 36: 213–224. https://doi.org/10.7872/crym/v36.iss2.2015.213</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Xu, L., Bao, D.F., Luo, Z.L., Su, X.J., Shen, H. & Su, H. (2020) Lignicolous freshwater ascomycota from Thailand: Phylogenetic and morphological characterisation of two new freshwater fungi: Tingoldiago hydei sp. nov. and T. clavata sp. nov. from Eastern Thailand. MycoKeys 65: 119–138. https://doi.org/10.3897/mycokeys.65.49769</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Yu, X.D., Zhang, S.N. & Liu, J.K. (2022) Morpho-Phylogenetic Evidence Reveals Novel Pleosporalean Taxa from Sichuan Province, China. Journal of Fungi 8: 720. https://doi.org/10.3390/jof8070720</span></span></span>
<p align="justify"><span style="color: #000000;"><span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, Y., Crous, P.W., Schoch, C.L. & Hyde, K.D. (2012) Pleosporales. Fungal Diversity 53: 1–221. https://doi.org/10.1007/s13225-011-0117-x</span></span></span>
<span style="font-family: Times New Roman, serif;"><span style="font-size: small;">Zhang, Y., Schoch, C.L., Fournier, J., Crous, P.W., de Gruyter, J., Woudenberg, J.H., Hirayama, K., Tanaka, K., Pointing, S.B., Spatafora, J.W. & Hyde, K.D. (2009) Multi-locus phylogeny of Pleosporales: a taxonomic, ecological and evolutionary re-evaluation. Studies in Mycology 64: 85–102. https://doi.org/10.3114/sim.2009.64.04</span></span>