Abstract
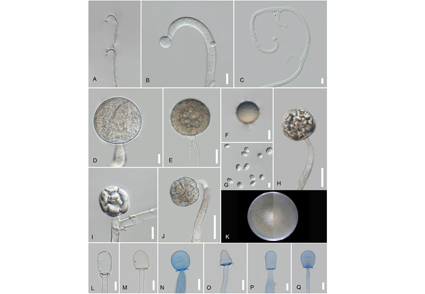
Backusella is morphologically and phylogenetically related to Mucor. Previously, due to various morphological similarities, distinction between these two genera was difficult and various Backusella species were classified within Mucor. However, in the last decade, with the advent of molecular phylogeny, the phylogenetic placement of the genus and its representative taxa was stabilised. In this study, a Backusella strain was isolated from soil samples in Thailand. A combination of morphology, phylogeny and physiology was used to characterize it. Phylogenetic analyses using the nuclear rDNA internal transcribed spacer (ITS1-5.8S-ITS2) and large subunit ribosomal ribonucleic acid (28S) genetic markers showed that the isolate is sister to Backusella gigacellularis. The new isolate is characterised by the production of simple or sympodially branched sporangiophores bearing a terminal sporangium. Columellae of sporangia are ellipsoidal, oblong to cylindrical, pyriform, and conical, often constricted in the centre. Sporangiola are globose to subglobose, often produced in short sympodially branched sporophores containing 4–8 sporangiospores each. The isolate grows on a relatively narrow temperature range of 15 °C–28 °C. The results of the three approaches indicated the novelty of the species. This is the first record of Backusella in Thailand.
References
Capella-Gutiérrez, S., Silla-Martínez, J.M. & Gabaldón, T. (2009) TrimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25: 1972–1973. https://doi.org/10.1093/bioinformatics/btp348
Crous, P.W., Carnegie, A.J., Wingfield, M.J., Sharma, R., Mughini, G., Noordeloos, M.E., Santini, A., Shouche, Y.S., Bezerra, J.D.P., Dima, B., Guarnaccia, V., Imrefi, I., Jurjevi?, Knapp, D.G., Kovács, G.M., Magistà, D., Perrone, G., Rämä, T., Rebriev, Y.A., Shivas, R.G., Singh, S.M., Souza-Motta, C.M., Thangavel, R., Adhapure, N.N., Alexandrova, A.V., Alfenas, A.C., Alfenas, R.F., Alvarado, P., Alves, A.L., Andrade, D.A., Andrade, J.P., Barbosa, R.N., Barili, A., Barnes, C.W., Baseia, I.G., Bellanger, J.M., Berlanas, C., Bessette, A.E., Bessette, A.R., Biketova, A.Y., Bomfim, F.S., Brandrud, T.E., Bransgrove, K., Brito, A.C.Q., Cano-Lira, J.F., Cantillo, T., Cavalcanti, A.D., Cheewangkoon, R., Chikowski, R.S., Conforto, C., Cordeiro, T.R.L., Craine, J.D., Cruz, R., Damm, U., De Oliveira, R.J.V., De Souza, J.T., De Souza, H.G., Dearnaley, J.D.W., Dimitrov, R.A., Dovana, F., Erhard, A., Esteve-Raventós, F., Félix, C.R., Ferisin, G., Fernandes, R.A., Ferreira, R.J., Ferro, L.O., Figueiredo, C.N., Frank, J.L., Freire, K.T.L.S., García, D., Gené, J., G?siorska, A., Gibertoni, T.B., Gondra, R.A.G., Gouliamova, D.E., Gramaje, D., Guard, F., Gusmão, L.F.P., Haitook, S., Hirooka, Y., Houbraken, J., Hubka, V., Inamdar, A., Iturriaga, T., Iturrieta-González, I., Jadan, M., Jiang, N., Justo, A., Kachalkin, A.V., Kapitonov, V.I., Karadelev, M., Karakehian, J., Kasuya, T., Kautmanová, I., Kruse, J., Kušan, I., Kuznetsova, T.A., Landell, M.F., Larsson, K.H., Lee, H.B., Lima, D.X., Lira, C.R.S., Machado, A.R., Madrid, H., Magalhães, O.M.C., Majerova, H., Malysheva, E.F., Mapperson, R.R., Marbach, P.A.S., Martín, M.P., Martín-Sanz, A., Mato?ec, N., McTaggart, A.R., Mello, J.F., Melo, R.F.R., Meši?, A., Michereff, S.J., Miller, A.N., Minoshima, A., Molinero-Ruiz, L., Morozova, O.V., Mosoh, D., Nabe, M., Naik, R., Nara, K., Nascimento, S.S., Neves, R.P., Olariaga, I., Oliveira, R.L., Oliveira, T.G.L., Ono, T., Ordoñez, M.E., Ottoni, A.M., Paiva, L.M., Pancorbo, F., Pant, B., Paw?owska, J., Peterson, S.W., Raudabaugh, D.B., Rodríguez-Andrade, E., Rubio, E., Rusevska, K., Santiago, A.L.C.M.A., Santos, A.C.S., Santos, C., Sazanova, N.A., Shah, S., Sharma, J., Silva, B.D.B., Siquier, J.L., Sonawane, M.S., Stchigel, A.M., Svetasheva, T., Tamakeaw, N., Telleria, M.T., Tiago, P.V., Tian, C.M., Tkal?ec, Z., Tomashevskaya, M.A., Truong, H.H., Vecherskii, M.V., Visagie, C.M., Vizzini, A., Yilmaz, N., Zmitrovich, I.V., Zvyagina, E.A., Boekhout, T., Kehlet, T., L?ss¸e, T. & Groenewald, J.Z. (2019) Fungal Planet description sheets: 868–950. Persoonia: Molecular Phylogeny and Evolution of Fungi 42: 291–473. https://doi.org/10.3767/persoonia.2019.42.11
Darriba, D., Taboada, G.L., Doallo, R. & Posada, D. (2012) jModelTest 2: more models, new heuristics and high-performance computing. Nature methods 9: 772. https://doi.org/10.1038/nmeth.2109
de Hoog, G.S. & Van Den Ende, A.H.G.G. (1998) Molecular diagnostics of clinical strains of filamentous Basidiomycetes: Molekulare diagnostik klinischer stämme filamentöser Basidiomyzeten. Mycoses 41: 183–189.
de Souza, J.I., Marano, A.V., Pires-Zottarelli, C.L.A., Chambergo, F.S. & Harakava, R. (2014) A new species of Backusella (Mucorales) from a Cerrado reserve in Southeast Brazil. Mycological Progress 13: 975–980. https://doi.org/10.1007/S11557-014-0981-3
Ellis, J.J. & Hesseltine, C.W. (1969) Two New Members of the Mucorales. Mycologia 61: 863–872. https://doi.org/10.1080/00275514.1969.12018810
Hoffmann, K., Paw?owska, J., Walther, G., Wrzosek, M., de Hoog, G.S., Benny, G.L., Kirk, P.M. & Voigt, K. (2013) The family structure of the Mucorales: a synoptic revision based on comprehensive multigene-genealogies. Persoonia 30: 57–76. https://doi.org/10.3767/003158513x666259
Huelsenbeck, J.P. & Ronquist, F. (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Hyde, K.D., Jeewon, R., Chen, Y.-J., Bhunjun, C.S., Calabon, M.S., Jiang, H.-B., Lin, C.-G., Norphanphoun, C., Sysouphanthong, P., Pem, D., Tibpromma, S., Zhang, Q., Doilom, M., Jayawardena, R.S., Liu, J.-K., Maharachchikumbura, S.S.N., Phukhamsakda, C., Phookamsak, R., Al-Sadi, A.M., Thongklang, N., Wang, Y., Gafforov, Y., Gareth Jones, E.B. & Lumyong, S. (2020) The numbers of fungi: is the descriptive curve flattening? Fungal Diversity 103: 219–271. https://doi.org/10.1007/s13225-020-00458-2
Hyde, K.D., Norphanphoun, C., Chen, J., Dissanayake, A.J., Doilom, M., Hongsanan, S., Jayawardena, R.S., Jeewon, R., Perera, R.H., Thongbai, B., Wanasinghe, D.N., Wisitrassameewong, K., Tibpromma, S. & Stadler, M. (2018) Thailand’s amazing diversity: up to 96% of fungi in northern Thailand may be novel. Fungal Diversity 93: 215–239. https://doi.org/10.1007/s13225-018-0415-7
Katoh, K. & Toh, H. (2008) Recent developments in the MAFFT multiple sequence alignment program. Briefings in Bioinformatics 9: 286–298. https://doi.org/10.1093/bib/bbn013
Kirk, P. (2012) Nomenclatural novelties Backusellaceae, Lentamycetaceae, Rhizopodaceae. Index Fungorum 11.
Kozlov, A.M., Darriba, D., Flouri, T., Morel, B. & Stamatakis, A. (2019) RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 35: 4453–4455. https://doi.org/10.1093/bioinformatics/btz305
Lima, D.X., Voigt, K., de Souza, C.A.F., de Oliveira, R.J.V., Souza-Motta, C.M. & Santiago, A.L.C.M. de A. (2016) Description of Backusella constricta sp. nov. (Mucorales, ex Zygomycota) from the Brazilian Atlantic Rainforest, including a key to species of Backusella. Phytotaxa 289: 59–68. https://doi.org/10.11646/phytotaxa.289.1.4
Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: 2010 Gateway Computing Environments Workshop (GCE). pp. 1–8.
Nguyen, T.T.T. & Lee, H.B. (2018) Isolation and characterization of three zygomycetous fungi in Korea: Backusella circina, Circinella muscae, and Mucor ramosissimus. Mycobiology 46: 317–327. https://doi.org/10.1080/12298093.2018.1538071
Nguyen, T.T.T., Voigt, K., Santiago, A.L.C.M. de A., Kirk, P.M. & Lee, H.B. (2021) Discovery of novel Backusella (Backusellaceae, Mucorales) isolated from invertebrates and toads in Cheongyang, Korea. Journal of Fungi 7: 513. https://doi.org/10.3390/jof7070513
Pidoplichko, N. & Milka, A. (1971) Atlas of mucoralean fungi. Acad. Sci. Ukranian SSR Kiev. pp. 188.
Senanayake, I., Rathnayaka, A., Marasinghe, D., Calabon, M., Lee, H., Hurdeal, V., Dissanayake, L., Wijesinghe, S., Nguyen, T., Goonasekara, I., Abeywickrama, P., Bhunjun, C., Jayawardena, R., Wanasinghe, D., Jeewon R, Bhat, D. & Xiang, M. (2020) Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11: 2678–2754. https://doi.org/10.5943/mycosphere/11/1/20
Trifinopoulos, J., Nguyen, L.-T., von Haeseler, A. & Minh, B.Q. (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Research 44: W232–W235. https://doi.org/10.1093/nar/gkw256
Urquhart, A.S., Douch, J.K., Heafield, T.A., Buddie, A.G. & Idnurm, A. (2021) Diversity of backusella (Mucoromycotina) in south-eastern australia revealed through polyphasic taxonomy. Persoonia: Molecular Phylogeny and Evolution of Fungi 46: 1–25. https://doi.org/10.3767/persoonia.2021.46.01
Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Walther, G., Paw?owska, J., Alastruey-Izquierdo, A., Wrzosek, M., Rodriguez-Tudela, J.L., Dolatabadi, S., Chakrabarti, A. & Hoog, G.S. de (2013) DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia: Molecular Phylogeny and Evolution of Fungi 30: 11–47. https://doi.org/10.3767/003158513x665070
Walther, G., Wagner, L. & Kurzai, O. (2019) Updates on the taxonomy of mucorales with an emphasis on clinically important taxa. Journal of Fungi 5. https://doi.org/10.3390/jof5040106
White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis, M.A., Gelfand, D.H., Sninsky, J.J. & White, T.J. (Eds.) PCR Protocols: a guide to methods and applications. Academic Press, San Diego, pp. 315–322.
Wijayawardene, N., Hyde, K., Al-Ani, L., Tedersoo, L., Haelewaters, D., K. C, R., Zhao, R.-L., Aptroot, A., Saxena, R., Tokarev, Y., Dai, D.-Q., Letcher, P., Stephenson, S., Dolatabadi, S., Lumbsch, T., Maharachchikumbura, S., Issi, I.V. & Madrid, H. (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11: 1160–1456. https://doi.org/10.5943/mycosphere/11/1/8