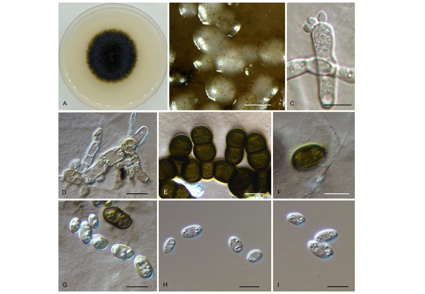
Abstract
During the surveys of air fungi in Beijing, China, a new ascomycetous black yeast-like species was isolated. Morphological and phylogenetical analyses of combined internal transcribed spacer (ITS) and the large ribosomal subunit (LSU) showed that it can be recognized as a new species of the genus Aureobasidium. Herein, Aureobasidium aerium sp. nov. is proposed and illustrated.
References
Arfi, Y., Marchand, C., Wartel, M. & Record, E. (2012) Fungal diversity in anoxic-sulfidic sediments in a mangrove soil. Fungal Ecology 5: 282–285. https://doi.org/10.1016/j.funeco.2011.09.004
Arzanlou, M. & Khodaei, S. (2012) Aureobasidium iranianum, a new species on bamboo from Iran. Mycosphere 3: 404–408. https://doi.org/10.5943/mycosphere/3/4/2
Cheng, J.H., Dai, S. & Ye, X.Y. (2016) Spatiotemporal heterogeneity of industrial pollution in China. China Economic Review 40: 179–191.
Crous, P.W., Summerell, B.A., Swart, L., Denman, S., Taylor, J.E., Bezuidenhout, C.M., Palm, M.E., Marincowitz, S. & Groenewald, J.Z. (2011) Fungal pathogens of Proteaceae. Persoonia 27: 20–45. https://doi.org/10.3767/003158511X606239
Cooke, W.B. (1963) A taxonomic study in the “black yeasts”. Mycopathologia et Mycologia Applicata 17: 1–43. https://doi.org/10.1007/BF02279261
Doyle, J.J. & Doyle, J.L. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 39–40.
Hermanides-Nijhof, E.J. (1977) Aureobasidium and allied genera. Studies in Mycology 15: 141–177.
Jiang, N., Liang, Y.M. & Tian, C.M. (2019) Aureobasidium pini sp. nov. from pine needle in China. Phytotaxa 402: 199–206. https://doi.org/10.11646/phytotaxa.402.4.3
Jiang, N., Liang, Y.M. & Tian, C.M. (2021) Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. Journal of Fungi 7: 64. https://doi.org/10.3390/jof7010064
Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. https://doi.org/10.1093/bib/bbx108
Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/msw054
Lee, D.H., Cho, S.E., Oh, J.Y., Cho, E.J. & Kwon, S. (2021) A novel species of Aureobasidium (Dothioraceae) recovered from Acer pseudosieboldianum in Korea. Journal of Asia-Pacific Biodiversity 14: 657–661. https://doi.org/10.1016/j.japb.2021.08.005
Li, Y., Chi, Z., Wang, G.Y., Wang, Z.P., Liu, G.L., Lee, C.F., Ma, Z.C. & Chi, Z.M. (2015) Taxonomy of Aureobasidium spp. and biosynthesis and regulation of their extracellular polymers. Critical Reviews in Microbiology 41: 228–237. https://doi.org/10.3109/1040841X.2013.826176
Ma, J., Xu, X., Zhao, C. & Yan, P. (2012) A review of atmospheric chemistry research in China: photochemical smog, haze pollution, and gas-aerosol interactions. Advances in Atmospheric Sciences 29: 1006–1026. https://doi.org/10.1007/s00376-012-1188-7
Nasr, S., Mohammadimehr, M., Vaghei, M.G., Amoozegar, M.A. & Fazeli, S.A.S. (2018) Aureobasidium mangrovei sp. nov., an ascomycetous species recovered from Hara protected forests in the Persian Gulf, Iran. Antonie van Leeuwenhoek 111: 1697–1705. https://doi.org/10.1007/s10482-018-1059-z
Nylander, J.A.A., Fredrik, R., Huelsenbeck, J.P. & Joséluis, N.A. (2004) Bayesian phylogenetic analysis of combined data. Systematic Biology 53: 47–67. https://doi.org/10.1080/10635150490264699
Peterson, S.W., Manitchotpisit, P. & Leathers, T.D. (2013) Aureobasidium thailandense sp. nov. isolated from leaves and wooden surfaces. International Journal of Systematic and Evolutionary Microbiology 63: 790–795. https://doi.org/10.1099/ijs.0.047613-0
Prasongsuk, S., Lotrakul, P., Ali, I., Bankeeree, W. & Punnapayak, H. (2018) The current status of Aureobasidium pullulans in biotechnology. Folia Microbiologica 63: 129–140. https://doi.org/10.1007/s12223-017-0561-4
Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Schoch, C.L., Shoemaker, R.A., Seifert, K.A., Hambleton, S., Spatafora, J.W. & Crous, P.W. (2006) A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1041–1052. https://doi.org/10.3852/mycologia.98.6.1041
Seifert, K.A. & Gams, W. (2011) The genera of Hyphomycetes–2011 update. Persoonia-Molecular Phylogeny and Evolution of Fungi 27: 119–129. https://doi.org/10.3767/003158511X617435
Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Swofford, D.L. (2003) PAUP*: Phylogenetic analysis using parsimony (* and other methods). Version 4.0b10. Sunderland, England, UK.
Thambugala, K.M., Ariyawansa, H.A., Li, Y.M., Boonmee, S., Hongsanan, S., Tian, Q., Singtripop, C., Bhat, D.J., Camporesi, E., Jayawardena, R., Liu, Z.Y., Xu, J.C., Chukeatirote, E. & Hyde, K.D. (2014) Dothideales. Fungal Diversity 68: 105–158. https://doi.org/10.1007/s13225-014-0303-8
Verkley, G.J., Starink-Willemse, M., van Iperen, A. & Abeln, E.C. (2004) Phylogenetic analyses of Septoria species based on the ITS and LSU-D2 regions of nuclear ribosomal DNA. Mycologia 96: 558–571. https://doi.org/10.1080/15572536.2005.11832954
Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Wei, X., Liu, G.L., Jia, S.L., Chi, Z., Hu, Z. & Chi, Z.M. (2021) Pullulan biosynthesis and its regulation in Aureobasidium spp. Carbohydrate Polymers 251: 117076. https://doi.org/10.1016/j.carbpol.2020.117076
White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322.
Wijayawardene, N.N., Hyde, K.D., Al-Ani, L.K.T. & et al. (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11: 1060–1456. https://doi.org/10.5943/mycosphere/11/1/8
Yurlova, N.A., Uijthof, J.M.J. & de Hoog, G.S. (1996) Distinction of species in Aureobasidium and related genera by PCR-ribotyping. Antonie van Leeuwenhoek 69: 323. https://doi.org/10.1007/BF00399621
Zalar, P., Gostin?ar, C., De Hoog, G.S., Urši?, V., Sudhadham, M. & Gunde-Cimerman, N. (2008) Redefinition of Aureobasidium pullulans and its varieties. Studies in Mycology 61: 21–38. https://doi.org/10.3114/sim.2008.61.02
Arzanlou, M. & Khodaei, S. (2012) Aureobasidium iranianum, a new species on bamboo from Iran. Mycosphere 3: 404–408. https://doi.org/10.5943/mycosphere/3/4/2
Cheng, J.H., Dai, S. & Ye, X.Y. (2016) Spatiotemporal heterogeneity of industrial pollution in China. China Economic Review 40: 179–191.
Crous, P.W., Summerell, B.A., Swart, L., Denman, S., Taylor, J.E., Bezuidenhout, C.M., Palm, M.E., Marincowitz, S. & Groenewald, J.Z. (2011) Fungal pathogens of Proteaceae. Persoonia 27: 20–45. https://doi.org/10.3767/003158511X606239
Cooke, W.B. (1963) A taxonomic study in the “black yeasts”. Mycopathologia et Mycologia Applicata 17: 1–43. https://doi.org/10.1007/BF02279261
Doyle, J.J. & Doyle, J.L. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 39–40.
Hermanides-Nijhof, E.J. (1977) Aureobasidium and allied genera. Studies in Mycology 15: 141–177.
Jiang, N., Liang, Y.M. & Tian, C.M. (2019) Aureobasidium pini sp. nov. from pine needle in China. Phytotaxa 402: 199–206. https://doi.org/10.11646/phytotaxa.402.4.3
Jiang, N., Liang, Y.M. & Tian, C.M. (2021) Identification and characterization of leaf-inhabiting fungi from Castanea plantations in China. Journal of Fungi 7: 64. https://doi.org/10.3390/jof7010064
Katoh, K., Rozewicki, J. & Yamada, K.D. (2019) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics 20: 1160–1166. https://doi.org/10.1093/bib/bbx108
Kumar, S., Stecher, G. & Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution 33: 1870–1874. https://doi.org/10.1093/molbev/msw054
Lee, D.H., Cho, S.E., Oh, J.Y., Cho, E.J. & Kwon, S. (2021) A novel species of Aureobasidium (Dothioraceae) recovered from Acer pseudosieboldianum in Korea. Journal of Asia-Pacific Biodiversity 14: 657–661. https://doi.org/10.1016/j.japb.2021.08.005
Li, Y., Chi, Z., Wang, G.Y., Wang, Z.P., Liu, G.L., Lee, C.F., Ma, Z.C. & Chi, Z.M. (2015) Taxonomy of Aureobasidium spp. and biosynthesis and regulation of their extracellular polymers. Critical Reviews in Microbiology 41: 228–237. https://doi.org/10.3109/1040841X.2013.826176
Ma, J., Xu, X., Zhao, C. & Yan, P. (2012) A review of atmospheric chemistry research in China: photochemical smog, haze pollution, and gas-aerosol interactions. Advances in Atmospheric Sciences 29: 1006–1026. https://doi.org/10.1007/s00376-012-1188-7
Nasr, S., Mohammadimehr, M., Vaghei, M.G., Amoozegar, M.A. & Fazeli, S.A.S. (2018) Aureobasidium mangrovei sp. nov., an ascomycetous species recovered from Hara protected forests in the Persian Gulf, Iran. Antonie van Leeuwenhoek 111: 1697–1705. https://doi.org/10.1007/s10482-018-1059-z
Nylander, J.A.A., Fredrik, R., Huelsenbeck, J.P. & Joséluis, N.A. (2004) Bayesian phylogenetic analysis of combined data. Systematic Biology 53: 47–67. https://doi.org/10.1080/10635150490264699
Peterson, S.W., Manitchotpisit, P. & Leathers, T.D. (2013) Aureobasidium thailandense sp. nov. isolated from leaves and wooden surfaces. International Journal of Systematic and Evolutionary Microbiology 63: 790–795. https://doi.org/10.1099/ijs.0.047613-0
Prasongsuk, S., Lotrakul, P., Ali, I., Bankeeree, W. & Punnapayak, H. (2018) The current status of Aureobasidium pullulans in biotechnology. Folia Microbiologica 63: 129–140. https://doi.org/10.1007/s12223-017-0561-4
Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574.
Schoch, C.L., Shoemaker, R.A., Seifert, K.A., Hambleton, S., Spatafora, J.W. & Crous, P.W. (2006) A multigene phylogeny of the Dothideomycetes using four nuclear loci. Mycologia 98: 1041–1052. https://doi.org/10.3852/mycologia.98.6.1041
Seifert, K.A. & Gams, W. (2011) The genera of Hyphomycetes–2011 update. Persoonia-Molecular Phylogeny and Evolution of Fungi 27: 119–129. https://doi.org/10.3767/003158511X617435
Stamatakis, A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Swofford, D.L. (2003) PAUP*: Phylogenetic analysis using parsimony (* and other methods). Version 4.0b10. Sunderland, England, UK.
Thambugala, K.M., Ariyawansa, H.A., Li, Y.M., Boonmee, S., Hongsanan, S., Tian, Q., Singtripop, C., Bhat, D.J., Camporesi, E., Jayawardena, R., Liu, Z.Y., Xu, J.C., Chukeatirote, E. & Hyde, K.D. (2014) Dothideales. Fungal Diversity 68: 105–158. https://doi.org/10.1007/s13225-014-0303-8
Verkley, G.J., Starink-Willemse, M., van Iperen, A. & Abeln, E.C. (2004) Phylogenetic analyses of Septoria species based on the ITS and LSU-D2 regions of nuclear ribosomal DNA. Mycologia 96: 558–571. https://doi.org/10.1080/15572536.2005.11832954
Vilgalys, R. & Hester, M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. https://doi.org/10.1128/jb.172.8.4238-4246.1990
Wei, X., Liu, G.L., Jia, S.L., Chi, Z., Hu, Z. & Chi, Z.M. (2021) Pullulan biosynthesis and its regulation in Aureobasidium spp. Carbohydrate Polymers 251: 117076. https://doi.org/10.1016/j.carbpol.2020.117076
White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR protocols: a guide to methods and applications 18: 315–322.
Wijayawardene, N.N., Hyde, K.D., Al-Ani, L.K.T. & et al. (2020) Outline of Fungi and fungus-like taxa. Mycosphere 11: 1060–1456. https://doi.org/10.5943/mycosphere/11/1/8
Yurlova, N.A., Uijthof, J.M.J. & de Hoog, G.S. (1996) Distinction of species in Aureobasidium and related genera by PCR-ribotyping. Antonie van Leeuwenhoek 69: 323. https://doi.org/10.1007/BF00399621
Zalar, P., Gostin?ar, C., De Hoog, G.S., Urši?, V., Sudhadham, M. & Gunde-Cimerman, N. (2008) Redefinition of Aureobasidium pullulans and its varieties. Studies in Mycology 61: 21–38. https://doi.org/10.3114/sim.2008.61.02