Abstract
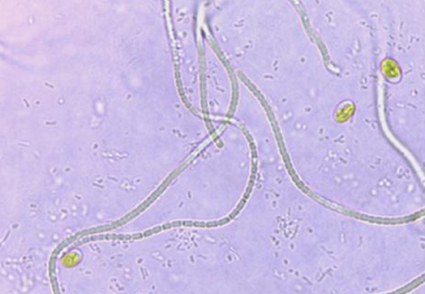
Two novel cyanobacteria (AP3 and AP3b) with thin cells and simple morphology were isolated from two islands of the Indian Sundarbans. The 16S rRNA phylogeny data revealed the distinct lineage of AP3b which was nearest to the clade incorporating the genus Oculatella and Tildeniella. Strain AP3 shared a common ancestor with the species Euryhalinema mangrovii. Additionally, the novel 16S rRNA gene sequences of strains AP3 and AP3b showed similarities about 98% and 93% respectively compared to those of established genera or species to which they were phylogenetically related. Furthermore, the folding patterns of semi-conservative structures like D1-D1’, Box-B and V2 helices of 16S-23S ITS region for both strains AP3 and AP3b displayed significant variations and uniqueness when compared with their respective reference strains (Euryhalinema mangrovii for AP3 and all the genera of Oculatellaceae for AP3b). Strain AP3 shared similar morphological features with its reference strain which confirmed its inter-species relationship. The diagnostic features of AP3b including the presence of necridic cells, aerotopes and a cluster-like growth pattern were found to be very contrasting. Altogether, these results substantiated the establishment of strain AP3b as a novel mono-specific genus named Aerofilum fasciculatum and strain AP3 as the second novel species under the genus Euryhalinema, referred to as Euryhalinema pallustris.
References
Boyer, S.L., Flechtner, V.R. & Johansen, J.R. (2001) Is the 16S–23S rRNA Internal Transcribed Spacer region a good tool for use in molecular systematics and population genetics? A case study in cyanobacteria. Molecular Biology and Evolution 18: 1057–1069. https://doi.org/10.1093/oxfordjournals.molbev.a003877
Chakraborty, S., Maruthanayagam, V., Achari, A., Mahansaria, R., Pramanik, A., Jaisankar, P. & Mukherjee, J. (2018) Oxynema aestuarii sp. nov. (Microcoleaceae) isolated from an Indian mangrove forest. Phytotaxa 374: 24–40. https://doi.org/10.11646/phytotaxa.374.1.2
Chakraborty, S., Maruthanayagam, V., Achari, A., Pramanik, A., Jaisankar, P. & Mukherjee, J. (2019) Euryhalinema mangrovii gen. nov., sp. nov. and Leptoelongatus litoralis gen. nov., sp. nov. (Leptolyngbyaceae) isolated from an Indian mangrove forest. Phytotaxa 422: 58–74. https://doi.org/10.11646/phytotaxa.422.1.4
Dadheech, P.K., Mahmoud, H., Kotut, K. & Krienitz, L. (2012) Haloleptolyngbya alcalis gen. et sp. nov., a new filamentous cyanobacterium from the soda lake Nakuru, Kenya. Hydrobiologia 691: 269–283. https://doi.org/10.1007/s10750-012-1080-6
Debnath, M., Singh, T. & Bhadury, P. (2017) New records of Cyanobacterial morphotypes with Leptolyngbya indica sp. nov. from terrestrial biofilms of the Lower Gangetic Plain, India. Phytotaxa 316: 101–120. https://doi.org/10.11646/phytotaxa.316.2.1
Demoulin, C.F., Lara, Y.J., Cornet, L., Francois, C., Baurain, D., Wilmotte, A. & Javaux, E.J. (2019) Cyanobacteria evolution: Insight from the fossil record. Free Radical Biology and Medicine 140: 206-223. https://doi.org/10.1016/j.freeradbiomed.2019.05.007
Drummond, A.J., Ho, S.Y.W., Philips, M.J. & Rambaut, A. (2006) Relaxed phylogenetics and dating with confidence. PLoS Biology 4: e88. https://doi.org/10.1371/journal.pbio.0040088
Erwin, P.M. & Thacker, R.W. (2008) Cryptic diversity of the symbiotic cyanobacterium Synechococcus spongiarum among sponge host. Molecular Ecology 17: 2937–2947. https://doi.org/10.1111/j.1365-294X.2008.03808.x.
Fiore, M.F., Sant’Anna, C.L., Azevedo, M.T.D., Komarek, J., Kastovsky, J., Sulek, J. & Lorenzi, A.S. (2007) The cyanobacterial genus Brasilonema, gen. nov., a molecular and phenotypic evaluation. Journal of Phycology 43: 789–798. https://doi.org/10.1111/j.1529-8817.2007.00376.x
Fuchsman, C.A., Collins, R.E., Rocap, G. & Brazelton, W.J. (2017) Effect of the environment on horizontal gene transfer between bacteria and archaea. PeerJ 5: e3865. https://doi.org/10.7717/peerj.3865
Gelman, A. & Rubin, D.B. (1992) Inference from iterative simulation using multiple sequences. Statistical Science 7: 457–472. https://doi.org/10.1214/ss/1177011136
Gonzalez-Resendiz, L., Johansen, J.R., Escobar-Sanchez, V., Segal- Kischinevzky, C., Jimenez-Garcia, L.F. & Leon-Tejera, H. (2018a) Two new species of Phyllonema (Rivulariaceae, Cyanobacteria) with an emendation of the genus. Journal of Phycology 54: 638–652. https://doi.org/10.1111/jpy.12769
Gonzalez-Resendiz, L., Johansen, J.R., Alba-Lois, L., Segal-Kischinevzky, C., Escobar-Sanchez, V., Jimenez Garcia, L.F., Hauer, T. & Leon-Tejera, H. (2018b) Nunduva, a new marine genus of Rivulariaceae (Nostocales, Cyanobacteria) from marine rocky shores. Fottea 18: 86–105. https://doi.org/10.5507/fot.2017.018
Gonzalez-Resendiz, L., Johansen, J.R., Leon-Tejera, H., Sanchez, L., Segal-Kischinevzky, C., Escobar-Sanchez, V. & Morales, M. (2019) A bridge too far in naming species: A total evidence approach does not support recognition of four species in Desertifilum (Cyanobacteria). Journal of Phycology 55: 898–911. https://doi.org/10.1111/jpy.12867
Iteman, I., Rippka, R., de Marsac, N.T. & Herdman, M. (2000) Comparison of conserved structural and regulatory domains within divergent 16S rRNA–23S rRNA spacer sequences of cyanobacteria. Microbiology 146: 1275–1286. https://doi.org/10.1099/00221287-146-6-1275
Johansen, J.R., Kovacik, L., Casamatta, D.A., Fu?iková, K. & Kaštovský, J. (2011) Utility of 16S-23S ITS sequence and secondary structure for recognition of intrageneric and intergeneric limits within cyanobacterial taxa: Leptolyngbya corticola sp. nov. (Pseudanabaenaceae, Cyanobacteria). Nova Hedwigia 92: 283–302. https://doi.org/10.1127/0029-5035/2011/0092-0283
Johansen, J.R., Mares, J., Pietrasiak, N., Bohunicka, M., Zima, J. Jr, Stenclova, L. & Hauer, T. (2017) Highly divergent 16S rRNA sequences in ribosomal operons of Scytonema hyalinum (Cyanobacteria). PLoS ONE 12: e0186393 https://doi.org/10.1371/journal.pone.0186393
Jung, P., Mikhailyuk, T., Emrich, D., Baumann, K., Dultz, S. & Budel, B. (2020) Shifting boundaries: Ecological and Geographical range extension based on three new species in the cyanobacterial genera Cyanocohniella, Oculatella and Aliterella. Journal of Phycology 56 (5): 1216–1231. [Early view] https://doi.org/10.1111/jpy.13025
Komarek, J. & Anagnostidis, K. (2005) Cyanoprokaryota. 2. Teil: Oscillatoriales. In: Büdel, B., Gärdner, G., Krienitz, L. & Schagerl, M. (Eds.) Süsswasserflora von Mitteleuropa. Elsevier, München, 759 pp.
Komarek, J., Kaštovský, J., Ventura, S., Turicchia, S. & Šmarda, J. (2009) The cyanobacterial genus Phormidesmis. Algological Studies 129: 41–59. https://doi.org/10.1127/1864-1318/2009/0129-0041
Komarek, J., Kastovsky, J., Mares, J. & Johansen, J.R. (2014) Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) using a polyphasic approach. Preslia 86: 295–335.
Lane, D.J. (1991) 16S/23S rRNA sequencing. In: Stackebrandt, E. & Goodfellow, M. (Eds.) Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley and Sons, pp. 115–175.
Larkin, M.A., Blackshields, G., Brown, N.P., Chenna, R., McGettigan, P.A., McWilliam, H., Valentin, F., Wallace, I.M., Wilm, A., Lopez, R., Thompson, J.D., Gibson, T.J. & Higgins, D.G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23: 2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Mai, T., Johansen, J.R., Pietrasiak, N., Bohunicka, M. & Martin, M.P. (2018) Revision of the Synechococcales (Cyanobacteria) through recognition of four families including Oculatellaceae fam. nov. and Trichocoleaceae fam. nov. and six new genera containing 14 species. Phytotaxa 365: 1–59. https://doi.org/10.11646/phytotaxa.365.1.1
Miller, M.A., Pfeiffer, W. & Schwartz, T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE) 2010: 1–8. https://doi.org/10.1109/GCE.2010.5676129
Neogi, S.B., Dey, M., Kabir, S.M.L., Masum, S.J.H., Kopprio, G., Yamasaki, S. & Lara, R. (2016) Sundarban mangroves: diversity, ecosystem services and climate change impacts. Asian Journal of Medical and Biological Research 2: 488–507. https://doi.org/10.3329/ajmbr.v2i4.30988
Nübel, U., Garcia-Pichel, F. & Muyzer, G. (1997) PCR primers to amplify 16S rRNA genes from cyanobacteria. Applied Environmental Microbiology 63: 3327–3332.
Osorio-Santos, K., Pietrasiak, N., Bohunická, M., Miscoe, L.H., Kovacik, L., Martin, M.P. & Johansen, J.R. (2014) Seven new species of Oculatella (Pseudanabaenales, Cyanobacteria) European Journal of Phycology 49: 450–470. https://doi.org/10.1080/09670262.2014.976843
Perkerson III, R.B., Johansen, J.R., Kovacik, L., Brand, J., Kastovsky, J. & Casamatta, D.A. (2011) A unique Pseudanabaenalean (cyanobacteria) genus Nodosilinea gen. nov. based on morphological and molecular data. Journal of Phycology 47: 1397–1412. https://doi.org/10.1111/j.1529-8817.2011.01077.x
Pietrasiak, N., Muhlsteinova, R., Siegesmund, M.A. & Johansen, J.R. (2014) Phylogenetic placement of Symplocastrum (Phormidiaceae) with a new combination S. californicum and two new species: S. flechtnerae and S. torsivum. Phycologia 53: 529–541. https://doi.org/10.2216/14-029.1
Pramanik, A., Sundararaman, M., Das, S., Ghosh, U. & Mukherjee, J. (2011) Isolation and characterization of cyanobacteria possessing antimicrobial activity from the Sundarbans, the world’s largest tidal mangrove forest. Journal of Phycology 47: 731–743. https://doi.org/10.1111/j.1529-8817.2011.01017.x
?eháková, K., Johansen, J.R., Bowen, M.B., Martin, M.P. & Sheil, C.A. (2014) Variation in secondary structure of the 16S rRNA molecule in cyanobacteria with implications for phylogenetic analysis. Fottea 14: 161–178. https://doi.org/10.5507/fot.2014.013
Rippka, R., Deruelles, J., Waterbury, J.B., Herdman, M. & Stanier, R.Y. (1979) Generic assignments, strain histories and properties of pure cultures of cyanobacteria. Journal of General Microbiology 111: 1–61. https://doi.org/10.1099/00221287-111-1-1
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M.A. & Huelsenbeck, J.P. (2012) MrBayes 3.2: effecient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. https://doi.org/10.1093/sysbio/sys029
Shalygin, S., Shalygina, R., Johansen, J.R., Pietrasiak, N., Berrendero Gomez, E., Bohunicka, M., Mares, J. & Sheil, C.A. (2017) Cyanomargarita gen. nov. (Nostocales, Cyanobacteria): convergent evolution resulting in a cryptic genus. Journal of Phycology 53: 762–777. https://doi.org/10.1111/jpy.12542
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. (2013) MEGA 6: Molecular Evolutionary Genetic Analysis Version 6.0. Molecular Biology and Evolution 30: 2725-2729. https://doi.org/10.1093/molbev/mst197
Tooming-Klunderud, A., Sogge, H., Rounge, T.B., Nederbragt, A.J., Lagesen, K., Glockner, G., Hayes, P.K., Rohrlack, T. & Jakobsen, K.J. (2013) From Green to Red: Horizontal Gene Transfer of the Phycoerythrin Gene cluster between Planktothrix strains. Applied and Environmental Microbiology 79: 6803-6812. https://doi.org/10.1128/AEM.01455-13
Turicchia, S., Ventura, S., Komárková, J. & Komárek, J. (2009) Taxonomic evaluation of cyanobacterial microflora from alkaline marshes of northern Belize: 2. Diversity of oscillatorialean genera. Nova Hedwigia 89: 65–200. https://doi.org/10.1127/0029-5035/2009/0089-0165
Vazquez-Martinez, J., Gutierrez-Villagomez, J.M., Fonesca-Garcia, C., Ramirez-Chavez, E., Mondragon-Sanchez, M.L., Partida- Martinez, L., Johansen, J.R. & Molina-Torres, J. (2018) Nodosilinea chupicuarensis sp. nov. (Leptolyngbyaceae, Synechococcales) a subaerial cyanobacterium isolated from a stone monument in central Mexico. Phytotaxa 334: 167–182. https://doi.org/10.11646/phytotaxa.334.2.6
Vinogradova, O., Mikhailyuk, T., Glaser, K., Holzinger, A. & Karsten, U. (2017) New species of Oculatella (Synechococcales, Cyanobacteria) from terrestrial habitats of Ukraine. Ukranian Botanical Journal 74: 509–520. https://doi.org/10.15407/ukrbotj74.06.509
Yarza, P., Yilmaz, P., Pruesse, E., Glöckner, F.O., Ludwig, W., Schleifer, K.H., Witman, W.B., Euzéby, J., Amann, R. & Rosselló-Móra, R. (2014) Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nature Reviews Microbiology 12: 635–645. https://doi.org/10.1038/nrmicro3330
Zammit, G. (2018) Systematics and biogeography of sciophilous cyanobacteria; an ecological and molecular description of Albertania skiophila (Leptolyngbyaceae) gen. & sp. nov. Phycologia 57: 481–491. https://doi.org/10.2216/17-125.1
Zammit, G., Billi, D. & Albertano, P. (2012) The subaerophytic cyanobacterium Oculatella subterranea (Oscillatoriales, Cyanophyceae) gen. et sp. nov.: a cytomorphological and molecular description. European Journal of Phycology 47: 341–354. https://doi.org/10.1080/09670262.2012.717106
Zhou, W., Ding, D., Yang, Q., Ahmad, M., Zhang, Y., Lin, X., Zhang, Y., Ling, J. & Dong, J. (2018) Marileptolyngbya sina gen. nov., sp. nov. and Salileptolyngbya diazotrophicum gen. nov., sp. nov. (Synechococcales, Cyanobacteria), species of cyanobacteria isolated from a marine ecosystem. Phytotaxa 383: 75–92. https://doi.org/10.11646/phytotaxa.383.1.4
Zuker, M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Research 31: 3406–3415. https://doi.org/10.1093/nar/gkg595