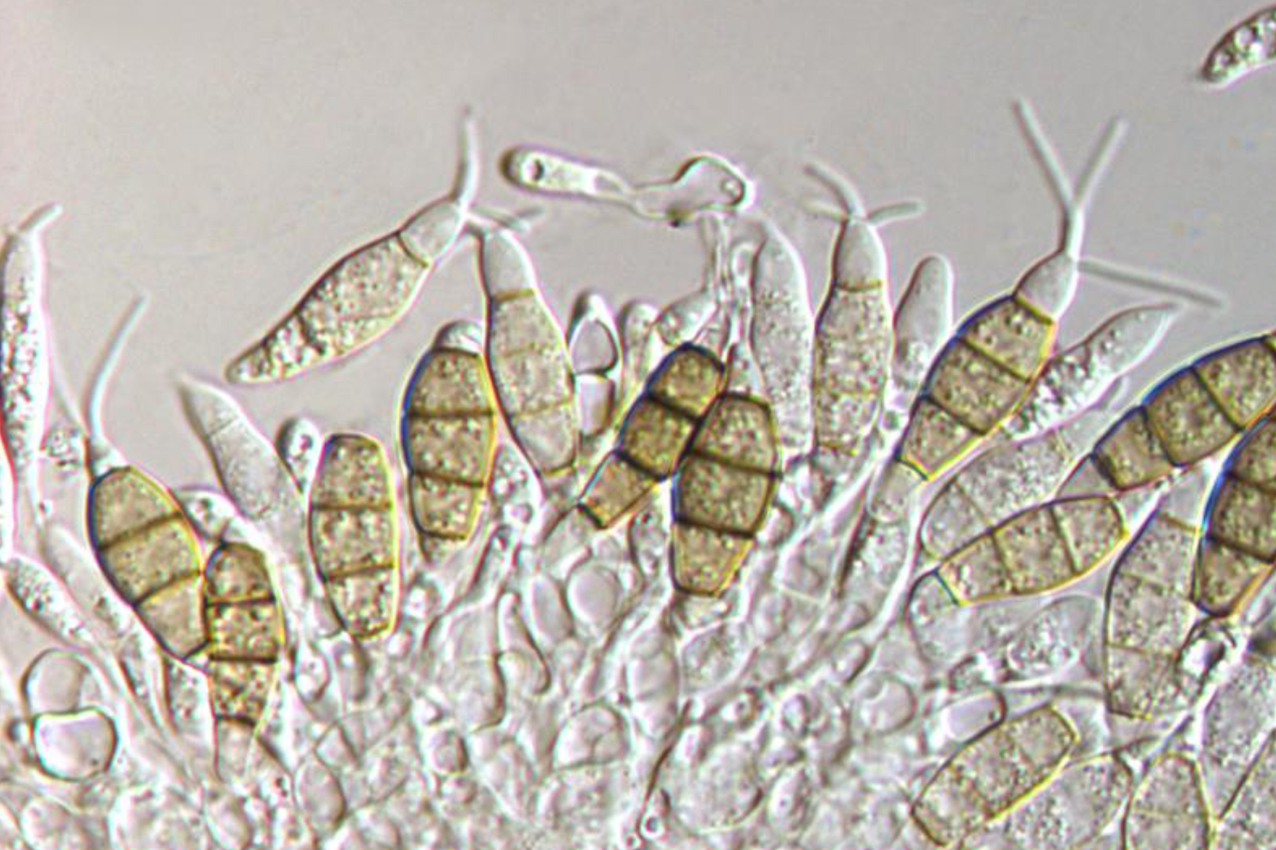
Abstract
Pestalotiopsis is a common genus worldwide and causes diseases on many plant hosts. During our investigation on plant diseases in Qinling Mountain in China, Pestalotiopsis species on needles of Abies fargesii was discovered. Based on the phylogeny of combined ITS, TEF and TUB genes, three Pestalotiopsis isolates from present study cluster into a distinct clade close to P. parva in the genus. However, conidia of Pestalotiopsis abietis are obviously larger than those from P. parva. Hence, Pestalotiopsis abietis sp. nov. is introduced herein.
References
Akinsanmi, O.A., Nisa, S., Jeff-Ego, O.S., Shivas, R.G. & Drenth, A. (2017) Dry flower disease of Macadamia in Australia caused by Neopestalotiopsis macadamiae sp. nov. and Pestalotiopsis macadamiae sp. nov. Plant disease 101 (1): 45–53. https://doi.org/10.1094/PDIS-05-16-0630-RE
Alves A, Crous P.W., Correia A. & Phillips A.J.L. (2008) Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity 28: 1–13.
Bate-Smith, E.C. & Metcalfe, C.R. (1957) Leuco-anthocyanins. 3. the nature and systematic distribution of tannins in dicotyledonous plants. Botanical Journal of the Linnean Society 55 (362): 669–705. https://doi.org/10.1111/j.1095-8339.1957.tb00030.x
Crous, P.W., Gams, W., Stalpers, J.A., Robert, V. & Stegehuis, G. (2004) Mycobank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22.
Dang, H., Zhang, Y., Zhang, Y., Zhang, K. & Zhang, Q. (2015) Variability and rapid response of subalpine fir (Abies fargesii) to climate warming at upper altitudinal limits in north-central China. Trees 29 (3): 785–795. https://doi.org/10.1007/s00468-015-1156-9
Das, R., Chutia, M., Das, K. & Jha, D.K. (2010) Factors affecting sporulation of Pestalotiopsis disseminata causing grey blight disease of Persea bombycina Kost., the primary food plant of muga silkworm. Crop Protection 29 (9): 963–968. https://doi.org/10.1016/j.cropro.2010.05.012
Doyle, J.J. & Doyle, J.L. (1990) Isolation of plant DNA from fresh tissue. Focus 12 (13): 39–40.
Felsenstein, J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Fu, L., Chen, T., Lang, K. & Hong T (2000) The Higher Plants of China, Volume 3. Qingdao Ocean Press, pp. 21–22.
Glass, N.L. & Donaldson, G. C. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61 (4): 1323–1330. https://doi.org/10.1128/AEM.61.4.1323-1330.1995
Geng, K., Zhang, B., Song, Y., Hyde, K.D., Kang, J.C. & Wang, Y. (2013) A new species of Pestalotiopsis from leaf spots of Licuala grandis from Hainan, China. Phytotaxa 88 (3): 49–54. https://doi.org/10.11646/phytotaxa.88.3.2
Guba, E.F. (1961) Monograph of Monochaetia and Pestalotia. Harvard University Press, Cambridge. https://doi.org/10.2307/3755860
Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59 (3): 307–321. https://doi.org/10.1093/sysbio/syq010
Hu, H., Jeewon, R., Zhou, D., Zhou, T. & Hyde, K. D. (2007) Phylogenetic diversity of endophytic Pestalotiopsis species in Pinus armandii and Ribes spp.: evidence from rDNA and ?-tubulin gene phylogenies. Fungal Diversity 24: 1–21.
Hyde, K.D., Hongsanan, S., Jeewon, R., Bhat, D.J., Mckenzie, E.H.C., Gareth, J.E.B., Phookamsak, R., Ariyawansa, H.A., Boonmee, S., Zhao, Q., Abdel-Aziz, F.A., Abdel-Waha, M.A., Banmai, S., Chomnunti, P., Cui, B.K., Daranagama, D., Das, K., Dayarathne, M.C., Silva, N.I.D., Dissanayake, A.J., Doilom, M., Ekanayaka, A.H., Gibertoni, T.B., Es-Neto, A.T.G., Huang, S.K., Jayasiri, S.C., Jayawardena, R.S., Konta, S., Lee, H.B., Li, W.J., Lin, C.G., Liu, J.K., Lu, Y.Z., Luo, Z.L., Manawasinghe, I.S., Manimohan, P., Mapook, A., Niskanen, T., Norphanphoun, C., Papizadeh, M., Perera, R.H., Phukhamsakda, C., Richter, C., Santiago, A.L.C.M.D.A., Drechsler-Santos, E.R., Senanayake, I.C. & Tanaka, K. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. https://doi.org/10.1007/s13225-016-0373-x
Jaklitsch, W.M., Gardiennet, A. & Voglmayr, H. (2016) Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 37: 82–105. https://doi.org/10.3767/003158516X690475
Katoh, K. & Toh, H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26 (15): 1899–1900. https://doi.org/10.1093/bioinformatics/btq224
Kohlmeyer, J. & Volkmann-Kohlmeyer, B. (2001) Fungi on Juncus roemerianus. 16. More new Coelomycetes, including Tetranacriella, gen. nov. Botanica Marina 44 (2): 147–156. https://doi.org/10.1515/BOT.2001.020
Li, G.J., Hyde, KD., Zhao, R.L., Hongsanan, S., Abdel-Aziz, F.A., Abdel-Wahab, M.A., Alvarado, P., Alves-Silva, G., Ammirati, J.F., Ariyawansa, H.A., Baghela, A., Bahkali, A.H., Beug, M., Bhat, D.J., Bojantchev, D., Boonpratuang, T., Bulgakov, T.S., Camporesi, E., Boro, M.C., Ceska, O., Chakraborty, D., Chen, J.J., Chethana, K.W.T., Chomnunti, P., Consiglio, G., Cui, B.K., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Das, K., Dayarathne, M.C., de Crop, E., de Oliveira, R.J.V., de Souza, C.A.F, de Souza, J.I., Dentinger, B.T.M., Dissanayake, A.J., Doilom, M., Drechsler-Santos, E.R., Ghobad-Nejhad, M., Gilmore, S.P. & Góes-Neto, A. (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal diversity 78 (1): 1–237. https://doi.org/10.1007/s13225-016-0366-9
Liu, F., Hou, L., Raza, M. & Cai, L. (2017) Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Scientific Reports 7 (1): 1–19. https://doi.org/10.1038/s41598-017-00972-5
Liu, J.K., Hyde, K.D., E.B.G. Jones, Ariyawansa, H.A., Bhat, J.D., Boonmee, S., Maharachchikumbura, S., McKenzie, E.H.C., Phookamsak, R., Phukhamsakda, C., Shenoy, B.D., Abdel-Wahab, M.A., Buyck, B., Chen, J., Chethana, K.W.T., Singtripop, C., Dai, D., Dai, Y., Daranagama, D.A., Dissanayake, A., Doilom, M., D’souza, M.J., Fan, X.L., Goonasekara, I.D., Hirayama, K., Hongsanan, S., Jayasiri, S.C., Jayawardena, R., Karunarathna, S.C., Li, W.J., Mapook, A., Norphanphoun, C., Pang, K.L., Perera, R.H., Peršoh, D., Pinruan, U., Senanayake, I., Somrithipol, S., Suetrong, S., Tanaka, K., Thambugala, K.M., Tian, Q., Tibpromma, S., Udayanga, D., Wijayawardene, N., Wanasinghe, D., Wisitrassameewong, K., Abdel-Aziz, F.A., Adam?ík, S., Bahkali, A.H., Boonyuen, N., Bulgakov, T., Callac, P., Chomnunti, P., Greiner, K., Hashimoto, A., Hofstetter, V., Kang, J.C., Lewis, D., Li, X.H., Liu, X.X., Liu, Z.Y., Matumura, M., Mortimer, P.E., Rambold, G., Randrianjohany, E., Sato, G., Indrasutdhi, V.S., Tian, C.M., Verbeken, A., von Brackel, W., Wang, Y., Wen, T.C., Xu, J.C., Yan, J.Y., Zhao, R.L. & Camporesi, E. (2015) Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. https://doi.org/10.1007/s13225-015-0324-y
Maharachchikumbura, S.S., Guo, L.D., Chukeatirote, E., Bahkali, A.H. & Hyde, K.D. (2011) Pestalotiopsis—morphology, phylogeny, biochemistry and diversity. Fungal Diversity 50 (1): 167–187. https://doi.org/10.1007/s13225-011-0125-x
Maharachchikumbura, S.S.N., Guo, L.D., Cai, L., Chukeatirote, E., Wu, W.P., Sun, X., Crous, P.W., Bhat, D.J., McKenzie, E.H.C., Bahkali, A.H. & Hyde, K.D. (2012) A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Diversity 56 (1): 95–129. https://doi.org/10.1007/s13225-012-0198-1
Maharachchikumbura, S.S., Zhang, Y.M., Wang, Y. & Hyde, K.D. (2013) Pestalotiopsis anacardiacearum sp. nov. Amphisphaeriaceae has an intricate relationship with Penicillaria jocosatrix, the mango tip borer. Phytotax 99 (2): 49–57. https://doi.org/10.11646/phytotaxa.99.2.1
Maharachchikumbura, S.S., Guo, L.D., Chukeatirote, E. & Hyde, K.D. (2014a) Improving the backbone tree for the genus Pestalotiopsis; addition of P. steyaertii and P. magna sp. nov. Mycological Progress 13 (3): 617–624. https://doi.org/10.1007/s11557-013-0944-0
Maharachchikumbura, S.S., Hyde, K.D., Groenewald, J.Z., Xu, J. & Crous, P.W. (2014b) Pestalotiopsis revisited. Studies in Mycology 79: 121–186. https://doi.org/10.1016/j.simyco.2014.09.005
O’Donnell, K. & Cigelnik, E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. https://doi.org/10.1006/mpev.1996.0376
Rannala, B., Yang, Z. (1996) Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. https://doi.org/10.1007/BF02338839
Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 (12): 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Silva, A.C., Diogo, E., Henriques, J., Ramos, A.P., Sandoval-Denis, M., Crous, P.W. & Bragança, H. (2020) Pestalotiopsis pini sp. nov., an emerging pathogen on stone pine (Pinus pinea L.). Forests 11 (8): 805. https://doi.org/10.3390/f11080805
Song, Y., Geng, K., Hyde, K.D., Zhao, W.S., Wei, J.G., Kang, J.C. & Wang, Y. (2013) Two new species of Pestalotiopsis from Southern China. Phytotaxa 126 (1): 22–32. https://doi.org/10.11646/phytotaxa.126.1.2
Song, Y., Maharachchikumbura, S.S., Jiang, Y.L., Hyde, K.D. & Wang, Y. (2014) Pestalotiopsis keteleeria sp. nov., isolated from Keteleeria pubescens in China. Chiang Mai Journal of Science 41 (4): 885–893.
Steyaert, R.L. (1949) Contribution a l’etude monographique de Pestalotia de Not. et Monochaetia Sacc. (Truncatella gen. nov. et Pestalotiopsis gen. nov.). Bulletin Jardin Botanique État Bruxelles 19: 285–354. https://doi.org/10.2307/3666710
Swofford, D.L. (2002) Phylogenetic analysis using parsimony (and other methods). Version 4.0 b10. Sunderland, Massachusetts: Sinauer Associates.
Tejesvi, M.V., Tamhankar, S.A., Kini, K.R., Rao, V.S. & Prakash, H.S. (2009) Phylogenetic analysis of endophytic Pestalotiopsis species from ethnopharmaceutically important medicinal trees. Fungal Diversity 38: 167–183.
Venkatasubbaiah, P., Grand, L.F. & Van Dyke, C.G. (1991) A new species of Pestalotiopsis on Oenothera. Mycologia 83 (4): 511–513. https://doi.org/10.1080/00275514.1991.12026042
Wei, J.G., Phan, C.K., Wang, L., Xu, T., Luo, J.T., Sun, X. & Guo, L.D. (2013) Pestalotiopsis yunnanensis sp. nov., an endophyte from Podocarpus macrophyllus (Podocarpaceae) based on morphology and ITS sequence data. Mycological Progress 12 (3): 563–568. https://doi.org/10.1007/s11557-012-0863-5
White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18 (1): 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Yasuda, F., Kobayashi, T., Watanabe, H. & Izawa, H. (2003) Addition of Pestalotiopsis spp. to leaf spot pathogens of Japanese persimmon. Journal of General Plant Pathology 69 (1): 29–32. https://doi.org/10.1007/s10327-002-0011-1
Zhang, Y., Maharachchikumbura, S.S., Mckenzie, E.H. & Hyde, K.D. (2012) A novel species of Pestalotiopsis causing leaf spots of Trachycarpus fortunei. Cryptogamie Mycologie 33 (3): 311–318. https://doi.org/10.7872/crym.v33.iss3.2012.311
Zhang, Y., Maharachchikumbura, S.S., Tian, Q. & Hyde, K.D. (2013) Pestalotiopsis species on ornamental plants in Yunnan Province, China. Sydowia 65 (1): 113–128.
Alves A, Crous P.W., Correia A. & Phillips A.J.L. (2008) Morphological and molecular data reveal cryptic speciation in Lasiodiplodia theobromae. Fungal Diversity 28: 1–13.
Bate-Smith, E.C. & Metcalfe, C.R. (1957) Leuco-anthocyanins. 3. the nature and systematic distribution of tannins in dicotyledonous plants. Botanical Journal of the Linnean Society 55 (362): 669–705. https://doi.org/10.1111/j.1095-8339.1957.tb00030.x
Crous, P.W., Gams, W., Stalpers, J.A., Robert, V. & Stegehuis, G. (2004) Mycobank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22.
Dang, H., Zhang, Y., Zhang, Y., Zhang, K. & Zhang, Q. (2015) Variability and rapid response of subalpine fir (Abies fargesii) to climate warming at upper altitudinal limits in north-central China. Trees 29 (3): 785–795. https://doi.org/10.1007/s00468-015-1156-9
Das, R., Chutia, M., Das, K. & Jha, D.K. (2010) Factors affecting sporulation of Pestalotiopsis disseminata causing grey blight disease of Persea bombycina Kost., the primary food plant of muga silkworm. Crop Protection 29 (9): 963–968. https://doi.org/10.1016/j.cropro.2010.05.012
Doyle, J.J. & Doyle, J.L. (1990) Isolation of plant DNA from fresh tissue. Focus 12 (13): 39–40.
Felsenstein, J. (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39: 783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Fu, L., Chen, T., Lang, K. & Hong T (2000) The Higher Plants of China, Volume 3. Qingdao Ocean Press, pp. 21–22.
Glass, N.L. & Donaldson, G. C. (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Applied and Environmental Microbiology 61 (4): 1323–1330. https://doi.org/10.1128/AEM.61.4.1323-1330.1995
Geng, K., Zhang, B., Song, Y., Hyde, K.D., Kang, J.C. & Wang, Y. (2013) A new species of Pestalotiopsis from leaf spots of Licuala grandis from Hainan, China. Phytotaxa 88 (3): 49–54. https://doi.org/10.11646/phytotaxa.88.3.2
Guba, E.F. (1961) Monograph of Monochaetia and Pestalotia. Harvard University Press, Cambridge. https://doi.org/10.2307/3755860
Guindon, S., Dufayard, J.F., Lefort, V., Anisimova, M., Hordijk, W. & Gascuel, O. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59 (3): 307–321. https://doi.org/10.1093/sysbio/syq010
Hu, H., Jeewon, R., Zhou, D., Zhou, T. & Hyde, K. D. (2007) Phylogenetic diversity of endophytic Pestalotiopsis species in Pinus armandii and Ribes spp.: evidence from rDNA and ?-tubulin gene phylogenies. Fungal Diversity 24: 1–21.
Hyde, K.D., Hongsanan, S., Jeewon, R., Bhat, D.J., Mckenzie, E.H.C., Gareth, J.E.B., Phookamsak, R., Ariyawansa, H.A., Boonmee, S., Zhao, Q., Abdel-Aziz, F.A., Abdel-Waha, M.A., Banmai, S., Chomnunti, P., Cui, B.K., Daranagama, D., Das, K., Dayarathne, M.C., Silva, N.I.D., Dissanayake, A.J., Doilom, M., Ekanayaka, A.H., Gibertoni, T.B., Es-Neto, A.T.G., Huang, S.K., Jayasiri, S.C., Jayawardena, R.S., Konta, S., Lee, H.B., Li, W.J., Lin, C.G., Liu, J.K., Lu, Y.Z., Luo, Z.L., Manawasinghe, I.S., Manimohan, P., Mapook, A., Niskanen, T., Norphanphoun, C., Papizadeh, M., Perera, R.H., Phukhamsakda, C., Richter, C., Santiago, A.L.C.M.D.A., Drechsler-Santos, E.R., Senanayake, I.C. & Tanaka, K. (2016) Fungal diversity notes 367–490: taxonomic and phylogenetic contributions to fungal taxa. Fungal Diversity 80: 1–270. https://doi.org/10.1007/s13225-016-0373-x
Jaklitsch, W.M., Gardiennet, A. & Voglmayr, H. (2016) Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia 37: 82–105. https://doi.org/10.3767/003158516X690475
Katoh, K. & Toh, H. (2010) Parallelization of the MAFFT multiple sequence alignment program. Bioinformatics 26 (15): 1899–1900. https://doi.org/10.1093/bioinformatics/btq224
Kohlmeyer, J. & Volkmann-Kohlmeyer, B. (2001) Fungi on Juncus roemerianus. 16. More new Coelomycetes, including Tetranacriella, gen. nov. Botanica Marina 44 (2): 147–156. https://doi.org/10.1515/BOT.2001.020
Li, G.J., Hyde, KD., Zhao, R.L., Hongsanan, S., Abdel-Aziz, F.A., Abdel-Wahab, M.A., Alvarado, P., Alves-Silva, G., Ammirati, J.F., Ariyawansa, H.A., Baghela, A., Bahkali, A.H., Beug, M., Bhat, D.J., Bojantchev, D., Boonpratuang, T., Bulgakov, T.S., Camporesi, E., Boro, M.C., Ceska, O., Chakraborty, D., Chen, J.J., Chethana, K.W.T., Chomnunti, P., Consiglio, G., Cui, B.K., Dai, D.Q., Dai, Y.C., Daranagama, D.A., Das, K., Dayarathne, M.C., de Crop, E., de Oliveira, R.J.V., de Souza, C.A.F, de Souza, J.I., Dentinger, B.T.M., Dissanayake, A.J., Doilom, M., Drechsler-Santos, E.R., Ghobad-Nejhad, M., Gilmore, S.P. & Góes-Neto, A. (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal diversity 78 (1): 1–237. https://doi.org/10.1007/s13225-016-0366-9
Liu, F., Hou, L., Raza, M. & Cai, L. (2017) Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Scientific Reports 7 (1): 1–19. https://doi.org/10.1038/s41598-017-00972-5
Liu, J.K., Hyde, K.D., E.B.G. Jones, Ariyawansa, H.A., Bhat, J.D., Boonmee, S., Maharachchikumbura, S., McKenzie, E.H.C., Phookamsak, R., Phukhamsakda, C., Shenoy, B.D., Abdel-Wahab, M.A., Buyck, B., Chen, J., Chethana, K.W.T., Singtripop, C., Dai, D., Dai, Y., Daranagama, D.A., Dissanayake, A., Doilom, M., D’souza, M.J., Fan, X.L., Goonasekara, I.D., Hirayama, K., Hongsanan, S., Jayasiri, S.C., Jayawardena, R., Karunarathna, S.C., Li, W.J., Mapook, A., Norphanphoun, C., Pang, K.L., Perera, R.H., Peršoh, D., Pinruan, U., Senanayake, I., Somrithipol, S., Suetrong, S., Tanaka, K., Thambugala, K.M., Tian, Q., Tibpromma, S., Udayanga, D., Wijayawardene, N., Wanasinghe, D., Wisitrassameewong, K., Abdel-Aziz, F.A., Adam?ík, S., Bahkali, A.H., Boonyuen, N., Bulgakov, T., Callac, P., Chomnunti, P., Greiner, K., Hashimoto, A., Hofstetter, V., Kang, J.C., Lewis, D., Li, X.H., Liu, X.X., Liu, Z.Y., Matumura, M., Mortimer, P.E., Rambold, G., Randrianjohany, E., Sato, G., Indrasutdhi, V.S., Tian, C.M., Verbeken, A., von Brackel, W., Wang, Y., Wen, T.C., Xu, J.C., Yan, J.Y., Zhao, R.L. & Camporesi, E. (2015) Fungal diversity notes 1–110: taxonomic and phylogenetic contributions to fungal species. Fungal Diversity 72: 1–197. https://doi.org/10.1007/s13225-015-0324-y
Maharachchikumbura, S.S., Guo, L.D., Chukeatirote, E., Bahkali, A.H. & Hyde, K.D. (2011) Pestalotiopsis—morphology, phylogeny, biochemistry and diversity. Fungal Diversity 50 (1): 167–187. https://doi.org/10.1007/s13225-011-0125-x
Maharachchikumbura, S.S.N., Guo, L.D., Cai, L., Chukeatirote, E., Wu, W.P., Sun, X., Crous, P.W., Bhat, D.J., McKenzie, E.H.C., Bahkali, A.H. & Hyde, K.D. (2012) A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Diversity 56 (1): 95–129. https://doi.org/10.1007/s13225-012-0198-1
Maharachchikumbura, S.S., Zhang, Y.M., Wang, Y. & Hyde, K.D. (2013) Pestalotiopsis anacardiacearum sp. nov. Amphisphaeriaceae has an intricate relationship with Penicillaria jocosatrix, the mango tip borer. Phytotax 99 (2): 49–57. https://doi.org/10.11646/phytotaxa.99.2.1
Maharachchikumbura, S.S., Guo, L.D., Chukeatirote, E. & Hyde, K.D. (2014a) Improving the backbone tree for the genus Pestalotiopsis; addition of P. steyaertii and P. magna sp. nov. Mycological Progress 13 (3): 617–624. https://doi.org/10.1007/s11557-013-0944-0
Maharachchikumbura, S.S., Hyde, K.D., Groenewald, J.Z., Xu, J. & Crous, P.W. (2014b) Pestalotiopsis revisited. Studies in Mycology 79: 121–186. https://doi.org/10.1016/j.simyco.2014.09.005
O’Donnell, K. & Cigelnik, E. (1997) Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Molecular Phylogenetics and Evolution 7: 103–116. https://doi.org/10.1006/mpev.1996.0376
Rannala, B., Yang, Z. (1996) Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. https://doi.org/10.1007/BF02338839
Ronquist, F. & Huelsenbeck, J.P. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19 (12): 1572–1574. https://doi.org/10.1093/bioinformatics/btg180
Silva, A.C., Diogo, E., Henriques, J., Ramos, A.P., Sandoval-Denis, M., Crous, P.W. & Bragança, H. (2020) Pestalotiopsis pini sp. nov., an emerging pathogen on stone pine (Pinus pinea L.). Forests 11 (8): 805. https://doi.org/10.3390/f11080805
Song, Y., Geng, K., Hyde, K.D., Zhao, W.S., Wei, J.G., Kang, J.C. & Wang, Y. (2013) Two new species of Pestalotiopsis from Southern China. Phytotaxa 126 (1): 22–32. https://doi.org/10.11646/phytotaxa.126.1.2
Song, Y., Maharachchikumbura, S.S., Jiang, Y.L., Hyde, K.D. & Wang, Y. (2014) Pestalotiopsis keteleeria sp. nov., isolated from Keteleeria pubescens in China. Chiang Mai Journal of Science 41 (4): 885–893.
Steyaert, R.L. (1949) Contribution a l’etude monographique de Pestalotia de Not. et Monochaetia Sacc. (Truncatella gen. nov. et Pestalotiopsis gen. nov.). Bulletin Jardin Botanique État Bruxelles 19: 285–354. https://doi.org/10.2307/3666710
Swofford, D.L. (2002) Phylogenetic analysis using parsimony (and other methods). Version 4.0 b10. Sunderland, Massachusetts: Sinauer Associates.
Tejesvi, M.V., Tamhankar, S.A., Kini, K.R., Rao, V.S. & Prakash, H.S. (2009) Phylogenetic analysis of endophytic Pestalotiopsis species from ethnopharmaceutically important medicinal trees. Fungal Diversity 38: 167–183.
Venkatasubbaiah, P., Grand, L.F. & Van Dyke, C.G. (1991) A new species of Pestalotiopsis on Oenothera. Mycologia 83 (4): 511–513. https://doi.org/10.1080/00275514.1991.12026042
Wei, J.G., Phan, C.K., Wang, L., Xu, T., Luo, J.T., Sun, X. & Guo, L.D. (2013) Pestalotiopsis yunnanensis sp. nov., an endophyte from Podocarpus macrophyllus (Podocarpaceae) based on morphology and ITS sequence data. Mycological Progress 12 (3): 563–568. https://doi.org/10.1007/s11557-012-0863-5
White, T.J., Bruns, T., Lee, S. & Taylor, J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: A Guide to Methods and Applications 18 (1): 315–322. https://doi.org/10.1016/B978-0-12-372180-8.50042-1
Yasuda, F., Kobayashi, T., Watanabe, H. & Izawa, H. (2003) Addition of Pestalotiopsis spp. to leaf spot pathogens of Japanese persimmon. Journal of General Plant Pathology 69 (1): 29–32. https://doi.org/10.1007/s10327-002-0011-1
Zhang, Y., Maharachchikumbura, S.S., Mckenzie, E.H. & Hyde, K.D. (2012) A novel species of Pestalotiopsis causing leaf spots of Trachycarpus fortunei. Cryptogamie Mycologie 33 (3): 311–318. https://doi.org/10.7872/crym.v33.iss3.2012.311
Zhang, Y., Maharachchikumbura, S.S., Tian, Q. & Hyde, K.D. (2013) Pestalotiopsis species on ornamental plants in Yunnan Province, China. Sydowia 65 (1): 113–128.